| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Review
Volume 4, December 2018, pages 88-98
Role of hydrophobicity in food peptide functionality and bioactivity
Caleb Acquah#, Elisa Di Stefano#, Chibuike C. Udenigwe*
School of Nutrition Sciences, University of Ottawa, Ottawa, Ontario, K1N 6N5, Canada
#These authors contributed equally.
*Corresponding author: Chibuike C. Udenigwe, School of Nutrition Sciences, University of Ottawa, Ottawa, Ontario, Canada, K1N 6N5. E-mail:
DOI: 10.31665/JFB.2018.4164
Received: November 23, 2018
Revised received & accepted: December 10, 2018
| Abstract | ▴Top |
Peptides are important compounds used in the development of functional biomaterials, functional foods and nutraceuticals. The functional and bioactive properties of peptides are directly linked to their structural features, including molecular size, presence or absence of charges, amino acid sequence, hydrophobicity, and hydrophilicity. The role of peptide structures in their bioactivities and functionalities is still emerging. Some bioactive peptides have undesirable taste, which can influence consumer interests in novel peptide-based food applications. In this review, we discussed the role of peptide hydrophobicity in their bioavailability, bioactivity, bitterness property, emulsion stability, aggregation and self-assembly for application in novel food formulations and nutraceutical/drug delivery.
Keywords: Bioactive Peptides; Structure; Hydrophobicity; Functionality; Bioactivity; Self-assembly; Bioavailability; Bitter taste
| 1. Introduction | ▴Top |
Research advances over the years have led to the development of several mainstream compounds for the treatment of diseases and for maintaining physiological homeostasis. However, the side-effects of some compounds can be burdensome (Fekete et al., 2015). Bioactive peptides have demonstrated several health-promoting effects that can be the basis of formulation of functional foods and nutraceuticals. This beneficial property has attracted the interest of consumers, food scientists, health professionals, and the food industry. Examples of physiological benefits derived from bioactive peptides include antioxidant, antihypertensive, antimicrobial, anti-inflammatory, anticalmodulin, anticancer, antithrombotic, and mineral binding activities (Agyei et al., 2017; Udenigwe 2014). The peptides discussed in this contribution are low molecular weight compounds generated via hydrolysis of food proteins by digestive, microbial, or plant proteases; in some cases, post-hydrolysis processes are used to isolate the active or functional molecules present in the complex hydrolysate mixture (Udenigwe and Aluko 2012).
The functional and bioactive properties of peptides are predicated on their structural features. The peptide structure encompasses properties such as molecular weight, net charge, amino acid sequence, hydrophobicity, and hydrophilicity. The type of enzyme used for proteolysis, the degree of hydrolysis, and processing conditions are known to influence the structure of peptides generated from proteins, which in turn affects the functional property, bitterness, biostability, cellular uptake, bioavailability, and bioactivity of peptides (Xie et al., 2015; Udenigwe and Aluko, 2012). Typically, the structure of food-derived bioactive peptides contains 2–20 amino acid residues, and is often predominated by hydrophobic chemical groups (Rani et al., 2018; Sánchez and Vázquez, 2017). Notably, the aforementioned structural features may act synergistically to promote the functionality and bioactivity of peptides (Yao et al., 2018). There is emerging interest in understanding the structure-property relationship of bioactive or functional peptides. This type of information is crucial in the design of potent or functionally useful peptides and in understanding their interaction with the food product and biological matrices. This contribution discusses the extant literature on the specific role of hydrophobicity in several functionalities and bioactivities of food protein-derived peptides.
| 2. Hydrophobicity of peptides | ▴Top |
Protein hydrolysis results in changes in the globular structure of the cleaved fragments, exposing amino acids of the hydrophobic regions, such as Phe, Tyr, Trp and Leu, which are typically hidden within the interior of the native protein structure (Al-Shamsi et al., 2017). Hydrophobicity plays a vital role in many functional and bioactive properties of food-derived peptides, as shown in Figure 1. For instance, the presence of aromatic amino acid residues at the C-terminus and hydrophobic amino acid residues at the N-terminus enhance the activity of peptides in inhibiting angiotensin I-converting enzyme (ACE) activity (Cheung et al., 2009; Majumder and Wu, 2009). Moreover, Xie et al., (2015) suggested that the hydrophobicity of casein-derived peptides could enhance their bioavailability and antioxidant properties, although a low digestive stability of the peptides was observed in simulated gastrointestinal fluids. Hydrophobic amino acids are also known to play a vital role in imparting a bitter taste to peptides (Seo et al., 2007). Owing to the importance of hydrophobicity in understanding the structure-property relationship, several amino acid hydrophobicity scales have been developed over the years. Table 1 shows the hydrophobicity scores of the 20 proteinogenic amino acids based on five hydrophobicity scales, with the Kyte and Doolittle scale being the most utilised in the literature. Hydrophobicity of peptide sequences can be calculated based on hydrophobicity of amino acid sidechains, using open access web-based bioinformatic tools (Table 2).
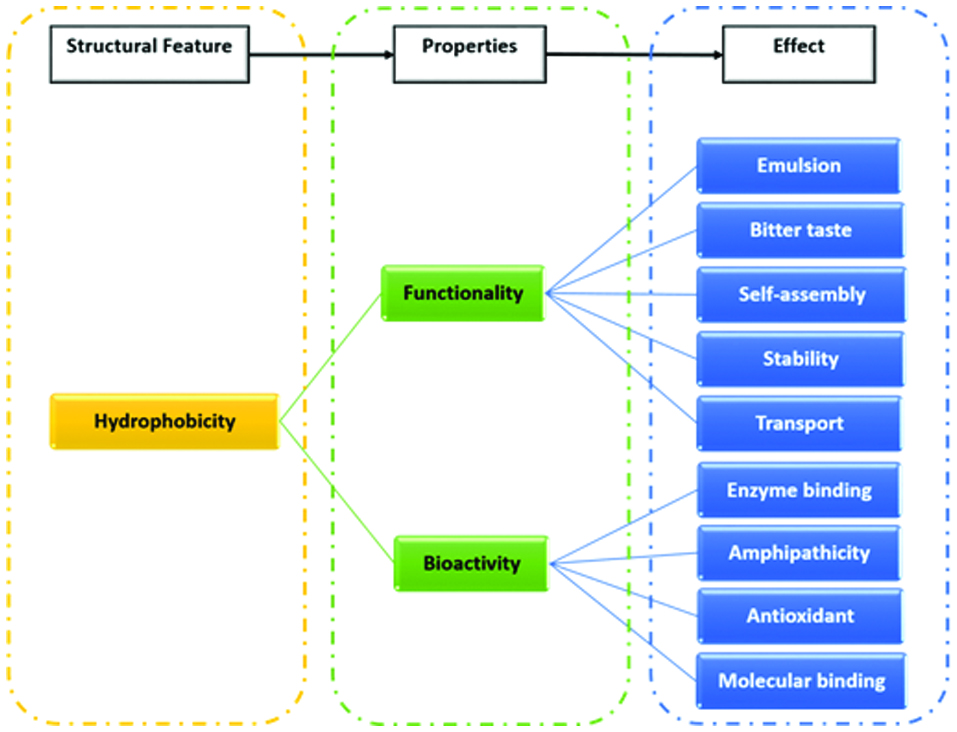 Click for large image | Figure 1. Role of hydrophobicity in several functionalities and health-related bioactivities of food protein-derived peptides. |
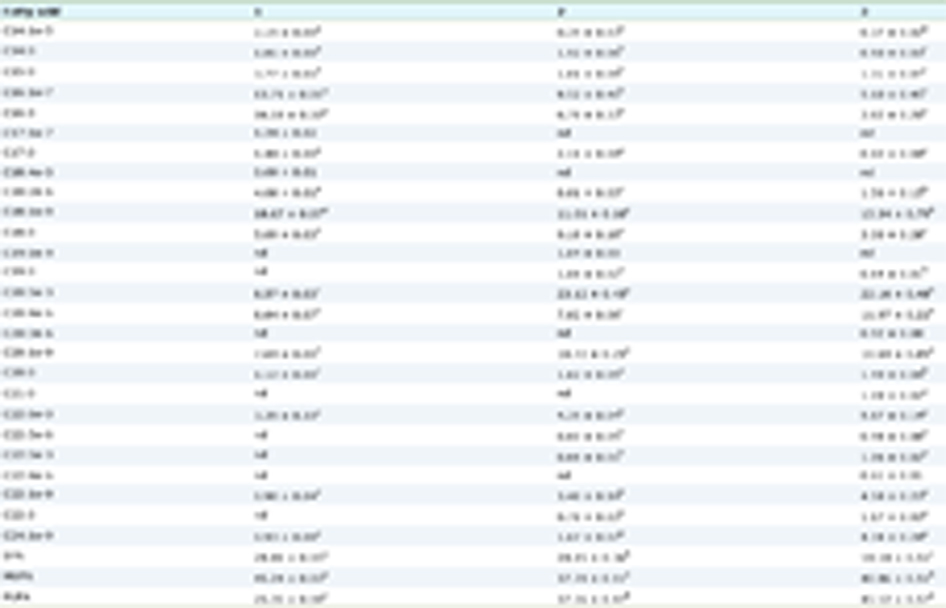 Click to view | Table 1. Amino acid hydrophobicity scales |
 Click to view | Table 2. Bioinformatic tools for calculating the hydrophobicity of peptides |
| 3. Role of hydrophobicity in peptide functionality | ▴Top |
3.1. Emulsion stability
Emulsions can exist in different forms including oil-in-water, water-in-oil, multiple emulsions (O-W-O or W-O-W), and nanoemulsions. Proteins have strong emulsion stabilizing effect owing to their high intrinsic surface activity, amphiphilicity, hydrophobicity, and capacity to quickly anchor at the oil-water or water-oil interface (Lam and Nickerson 2013; Al-Shamsi et al., 2018). Strong emulsifiers can stabilize small droplet sizes and this property positively correlates with the surface hydrophobicity of proteins (Chen et al., 2016). Thus, protein hydrolysis can release peptides with exposed hydrophobic regions, which would promote their emulsion stabilizing effect (Figure 2). For instance, Qi et al., (1997) demonstrated that hydrolysis of soy protein isolates with pancreatin resulted in increased surface hydrophobicity and emulsifying capacity when compared to the native soy protein isolates. Notably, modification of proteins by limited hydrolysis leads to a high interfacial activity and thus an initial rapid foaming and emulsifying capacity (Hall et al., 2017). However, the foaming and emulsion stabilizing effect of protein hydrolysates could also decrease over time due to the reduction in molecular size of the polypeptides and changes in hydrophobicity during extensive hydrolysis, which affects the strength of the interfacial layer and formation of networks (Hall et al., 2017; Miñones Conde and Rodríguez Patino, 2007). Modification of proteins by controlling the degree of hydrolysis can result in their structural rearrangement, increase in solubility, reduction of molecular size, exposure of hydrophobic amino acids, and increase in surface hydrophobicity to enhance the emulsifying stabilizing capacity of peptides (Lam and Nickerson, 2013; Tsumura, 2009).
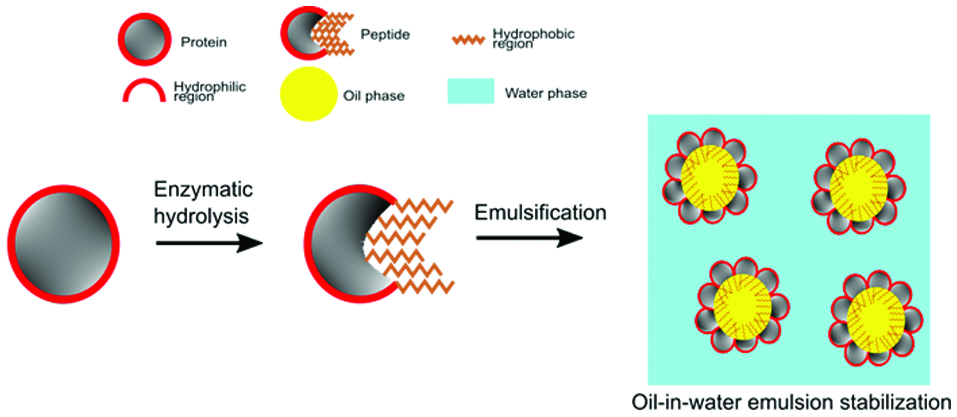 Click for large image | Figure 2. Enzymatic hydrolysis in producing hydrophobic peptides for stabilizing emulsions. |
An improvement in emulsifying capacity has also been reported by Chen et al. (2016) after incorporating high-pressure microfluidization pre-treatment prior to hydrolysis of soy protein isolate with pancreatin. It was suggested that high-pressure microfluidization of soy protein isolates resulted in structural rearrangement that enhanced the accessibility of particular subunits, such as α′-7S, A-11S and B-11S, to the enzyme. This resulted in a smaller mean droplet size, increased protein solubility, reduced molecular weight, increased surface hydrophobicity, and improved emulsion stability.
3.2. Self-assembly and aggregation
Self-assembly is a spontaneous process by which molecular units organise into ordered structures through intermolecular and intramolecular interactions. The spontaneous process is regulated by balancing the attractive and repulsive forces existing between and within the molecules (Mandal et al., 2014). Molecules that undergo self-assembly are generally amphiphilic and thus contain both hydrophobic and hydrophilic moieties. Peptides can be amphiphilic and thus influenced to undergo self-assembly by factors such as concentration, temperature, pH, and ionic strength of their environment. Figure 3 shows several applications of peptide self-assembly. The common motifs involved in peptide self-assembly are the association of β-strands and α-helices (Doll et al., 2013; Deming, 2005). Molecular self-assembly occurs above the critical aggregation concentration of a compound and is thought to be driven mostly by non-covalent forces such as electrostatic interactions, van der Waals, π-stacking and hydrogen bonding, with the latter being the most influential (Hutchinson et al., 2017). Ganesan and Matysiak (2016) demonstrated that backbone interpeptide dipolar interactions contributed significantly in the aggregation of fibril-like peptides, and that the hydrophobic-hydrophilic interface enhanced the order and rate of aggregation. Moreover, Kim and Hecht (2006) reported that hydrophobic amino acid residues present at the C-terminal of amyloid β peptide contributed in triggering aggregation of the peptide. The process of peptide self-assembly and aggregation is complex. Several models have been used to predict aggregation mechanisms due to the lack of suitable experimental techniques to unravel their early formation at the atomic level (Meli et al., 2008).
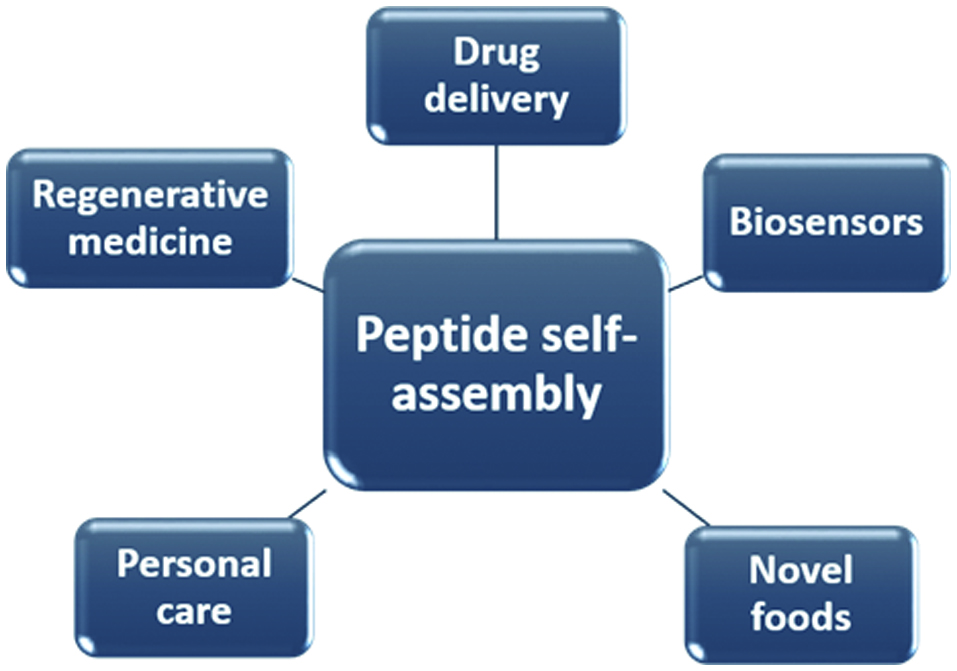 Click for large image | Figure 3. Various applications of peptide self-assembly. |
During assembly, hydrogen bonding existing between backbone structures causes peptide monomers to align longitudinally into β-sheets. The inter-β-sheets interactions on the side chains of peptide molecules control the slow, lateral packing of β-sheets (Zhou et al., 2016). All amino acids, with the exception of glycine, contain a chiral carbon atom in the L configuration. The intrinsic chirality of amino acids causes a twist in the lateral packing of β-sheets, which affects the final morphology such as nanofibers, nanovesicles, nanomicelles, nanofibrils and nanotubes of the self-assembled peptides (Cui et al., 2009). Notably, the stronger the inter-β-sheet interactions on the side chains of peptides, the better the lateral packing. Peptides are good candidates for developing self-assembled nanostructures for biological applications due to their structural and functional diversity, and biocomptability (Mandal et al., 2014). Further studies are needed to advance current knowledge in food peptide aggregation and self-assembly, specifically in identifying the driving forces and formation mechanisms, and adapting the peptide products for food and biomedical applications.
3.3. Biostability and transport across intestinal cells
Biostability and cellular uptake of peptides are highly influenced by their molecular size, hydrophobicity, and net charge (Chua et al., 2004; Wang and Li, 2017). Although bioactive peptides are susceptible to degradation by gastrointestinal proteases and serum peptidases, numerous in vitro studies in human intestinal (Caco-2) cell monolayers have demonstrated that the transport of some peptides can occur successfully without altering their bioactivities (Fu et al., 2016; Fernández-Tomé et al., 2018; Vij et al., 2016). Resistance to peptidases during intestinal transport is thought to be dependent on the specific peptide and positioning of their amino acid residues (Fu et al., 2016). According to Shimizu et al., (1997), N-terminal amino acid residues are more prone to intestinal peptidase degradation compared to C-terminal residues, and this may influence the bioavailability of peptides in the gut. Ohsawa et al., (2008) suggested that the presence of Pro residues could enhance the resistance of antihypertensive peptides to peptidase activities. The positioning of hydrophobic aromatic amino acids, Tyr, Phe and Trp, at the C-terminal could also enhance the resistance of peptides to intestinal peptidases (Bejjani and Wu 2013). In a recent study, Yang et al., (2017) demonstrated that the structural stability, transport across Caco-2 cell monolayers and bioactivity of Leu-Tyr, Arg-Ala-Leu-Pro and Thr-Phe may be dependent on hydrophobicity. They found that Thr-Phe was highly prone to human intestinal brush border amino peptidases followed by Arg-Ala-Leu-Pro, whereas hydrophobic Leu-Tyr retained its structural integrity and activity in inhibiting ACE and renin, after peptidase treatment. Furthermore, lipophilicity of peptides (i.e., hydrophobicity and hydrogen bonding potential) is crucial in transcellular intestinal transport of peptides, by facilitating their interaction with cell membranes (Pauletti et al., 1997). Peptide modifications (e.g. cyclization) that increase hydrophobicity and decrease hydrogen bonding potential are considered effective approaches for enhancing oral bioavailability of peptide drugs (Pauletti et al., 1997). Hydrophobicity can also influence the bioaccessibility of peptides in the gastrointestinal tract, but further studies are needed to validate these relationships for food protein-derived peptides.
3.4. Hydrophobicity-taste relationship
Factors determining the bitterness of peptides include degree of hydrolysis, processing conditions, type of protease used in protein hydrolysis, average hydrophobicity value, and sequence of hydrophobic amino acid residues (Murray et al., 2018; Seo et al., 2007; Iwaniak et al., 2018). Other factors such as modification by acetylation or esterification of the carboxyl and amino groups could also exacerbate the undesirable organoleptic properties of peptides (Kohl et al., 2012). Bitterness property of functional foods can be evident when there is a disproportionate concentration of bitter peptides amongst other food bioactive constituents. Heinz Ney (1979) demonstrated that peptide bitterness is mainly attributed to the presence of hydrophobic amino acid residues, and proposed the Q rule. The Q value specifies average hydrophobicity based on individual free energies of the amino acid residues, as shown in the following equation:
Variations in the length of peptide sequence stem from the use of proteases with different specificities in cleaving peptide bonds. Humiski and Aluko (2007) found that Alcalase and Flavourzyme, out of five different proteases, produce more bitter peptides due to their specificity for hydrophobic amino acid residues, whereas papain and α-chymotrypsin had the most desirable taste scores. There was no observed correlation between molecular size and bitterness scores of the peptides from pea protein hydrolysates (Humiski and Aluko, 2007). Also, total content of hydrophobic free amino acids, such as Val, Ile, Phe, Trp, Leu and Tyr, were observed to increase with bitterness when Alcalase was used to produce cricket protein hydrolysates (Hall et al., 2017). This finding is in agreement with that of Seo et al. (2007) who observed that Alcalase produced the highest bitter score after soy protein hydrolysis, followed by Neutrase, Protamex, papain, bromelain, and Flavourzyme.
Recently, a chemometric analysis of dipeptides and tripeptides showed that bulky and branched sidechains of Leu, Ile, Val, Tyr, Phe and Trp, when present at the C-terminal, increase the bitterness of peptides (Iwaniak et al., 2018). Using C6-glioma cells that express bitter taste receptor (T2R)1, Upadhyaya et al. (2010) demonstrated that the highly hydrophobic tripeptide Phe-Phe-Phe is capable of activating the receptor based on observed changes in intracellular calcium levels. The role of hydrophobic amino acid residues in bitterness is thought to be related to their binding of the human taste G-protein coupled receptors (GPCR), and activation of associated sensory signalling processes. However, Zhang et al., (2018) recently reported that, although hydrophobicity enabled the bioaccessibility to target organs, two beef-derived peptides, Ala-Gly-Asp-Asp-Ala-Pro-Arg-Ala-Val-Phe and Glu-Thr-Ser-Ala-Arg-His-Leu, suppressed the activation of quinine-dependent T2R4 expressing cells due to their high hydrophilicity (>50%). This finding underscores the possible contribution of other structural and environmental factors in determining molecular interactions and biological effects.
| 4. Role of hydrophobicity in peptide bioactivity | ▴Top |
4.1. Enzyme binding and inhibition
Due to similarity with the structure with human regulatory peptide motifs, several food-derived peptides are able to interact with enzymes and receptors involved in the regulation of inflammatory process, blood glucose level, and blood pressure, such as ACE, dipeptidyl peptidase IV (DPP-IV), renin, and α-glucosidase. In many instances, as discussed below, peptide hydrophobicity is thought to play crucial roles in stabilizing these molecular interactions and promoting bioactivity.
4.1.1. Alpha-glucosidase inhibition
Alpha (α)-glucosidase, a human enzyme located in the epithelial mucosa of the small intestine, is responsible for the hydrolysis and release of terminal, non-reducing glucose moieties from food-derived disaccharides. Due to the enzyme’s central role in regulating postprandial glucose release, α-glucosidase inhibitors have been developed for type 2 diabetes management. The presence of hydrophobic amino acids Pro, Met and Ala, close to the C-terminal of the peptide, was observed to contribute in promoting the inhibitory activity (Kang et al., 2013; Mojica et al., 2017). Molecular docking studies also revealed that hydrophobic amino acid residues of peptides predominantly interact with residues in the active site of α-glucosidase (Mojica and de Mejía, 2016; Di Stefano et al., 2018). In addition, QSAR models of α-glucosidase inhibitors have confirmed the importance of molecular hydrophobicity in their bioactivity, and also showed that a balance between hydrophobic and hydrophilic residues is crucial for the formation of H-bonds and electrostatic interactions involved in the enzyme-inhibitor complexation (Moorthy et al., 2011). Besides the involvement of hydrophobic amino acid residues, the mechanisms involved in the α-glucosidase inhibitory activity of peptides still need to be elucidated.
4.1.2. Angiotensin I-converting enzyme inhibition
The renin-angiotensin system is the major pathway involved in blood pressure control, where renin is responsible for the conversion of angiotensinogen into angiotensin I (rate limiting step), and ACE subsequently converts angiotensin I into the strong vasoconstrictor angiotensin II, while inactivating the vasodilator, bradykinin. Because of its crucial involvement in blood pressure regulation, ACE inhibitors are currently used as drugs for treating hypertension. When considering the ACE inhibitory activity of peptides, the presence of hydrophobic amino acids Pro, Phe, Ile, Val, close to the C-terminus of peptides promotes their binding to ACE and ultimately intensifies enzyme inhibition (Cheung et al., 1980; Fu et al., 2016). For instance, the most active ACE inhibiting peptides from enzymatic hydrolysis of egg white protein were hydrophobic (Phe-Arg-Ala-Asp-His-Pro-Phe-Leu, IC50 3.2 µM; Tyr-Ala-Glu-Glu-Arg-Tyr-Pro-Ile-Leu, IC50 4.7 µM; and Arg-Ala-Asp-His-Pro-Phe-Leu, IC50 6.2 µM), and that from digested squid skin collagen was a proline-rich decapeptide with an IC50 of 48 µM (Miguel et al., 2004; Alemán et al., 2013). ACE inhibiting peptides isolated from digested pork meat (Arg-Pro-Arg and Pro-Thr-Pro-Val-Pro) and from Spanish dry-cured ham (Ala-Ala-Ala-Thr-Pro) also showed similar hydrophobicity features (Escudero et al., 2012, 2013). The crucial role of hydrophobic amino acids was demonstrated by Saiga et al. (2006) with the ACE inhibitory polypeptide Gly-Phe-Hyp-Gly-Thr-Hyp-Gly-Leu-Hyp-Gly-Phe isolated from digested chicken breast muscle, which had its IC50 value increased (i.e. decrease in activity) from 42 µM to 52.5 µM by the removal of the C-terminal Phe residue using ACE. Interestingly, other fragments of the peptide with C-terminal Phe (i.e. Hyp-Gly-Leu-Hyp-Gly-Phe and Hyp-Gly-Thr-Hyp-Gly-Leu-Hyp-Gly-Phe) exhibited relatively higher ACE inhibitory activities with IC50 values of 10 and 19 µM, respectively. These structure-activity relationships were established based on in vitro studies. It would be beneficial to confirm the role of hydrophobicity in lowering of blood pressure when the bioactive peptides are present in complex food and physiological environments.
4.1.3. Dipeptidyl peptidase-IV inhibition
DPP-IV is a metabolic enzyme that selectively converts the incretin hormones, gastric inhibitory peptide and glucagon-like peptide-1, into their inactive forms, thereby largely influencing postprandial insulin release. Because of its detrimental activity towards postprandial glucose regulation, DPP-IV inhibitory drugs called gliptins have been developed as a therapeutic strategy for type 2 diabetes mellitus (Liu et al., 2017). DPP-IV contains a hydrophobic site S1 with Tyr631, Val656, Trp659, Tyr666 and Val711 residues, and a charged site S2 with Arg125, Glu205, Glu206, Phe357, Ser209 and Arg358 residues (Power et al., 2014). Several studies using bioinformatics, in vitro and animal models have demonstrated that food proteins contain DPP-IV inhibiting peptides within their primary structures (Liu et al., 2017; Udenigwe et al., 2013). These peptides can be released using methods such as enzymatic hydrolysis and fermentation. DPP-IV inhibiting compounds exert their bioactivity by interacting with the enzyme active site through salt bridges, hydrophobic interactions, and H-bonds. Notably, the interaction with drug inhibitors mostly occurs at the Arg125, Glu205, Glu206, Tyr547, Tyr662, Tyr666, Ser630 and Phe357 residues of the enzyme, whereas the Glu205, Glu206 and Ser630 residues interact with the N-terminus hydrophobic region of some peptides (Arulmozhiraja et al., 2016). In general, food-derived peptides were observed to interact with both the hydrophobic S1 and charged S2 pockets of DPP-IV. For instance, Liu et al., (2017) reported that DPP-IV inhibiting peptides LAPSTM (IC50 140 µM), FAGDDAPR (IC50 168 µM) and FAGDDAPRA (IC50 393 µM) interacted with regions of the enzyme through H-bonding and π-π interactions. The presence of hydrophobic amino acids at the N-terminus of peptides is a peculiar feature of identified DPP-IV inhibiting peptides. In addition to hydrophobicity, the penultimate N-terminal position appears to play a crucial role in enhancing or decreasing the bioactivity. Harnedy et al. (2015) reported that the DPP-IV inhibitory activity of peptides ILAP and LLAP (IC50 of 43.4 and 53.7 μM, respectively) dropped to insignificant values when the penultimate N-terminal Leu was replaced with Ile. Although many studies have identified food-derived DPP-IV inhibiting peptides, there is still a dearth of information on the role of the generally hydrophobic characters of the peptide in their transport through the intestinal epithelium and retention of their bioactivity once absorbed and distributed in the body.
4.2. Amphipathicity in antimicrobial activity
Cationic antimicrobial peptides (CAMPs) are net positively charged peptides of 12-50 amino acid residues, with approximately half of the structure being hydrophobic. CAMPs are naturally present in many organisms, where they play a non-specific defensive role against microbial infections (Hancock, 2001). The high content of lysine and arginine residues determines the cationic nature of the peptides. A combination of cationic patches and hydrophobic cores of CAMPs result in an amphipathicity and rearrangement of the peptides into specific 2D or 3D structures, which is crucial for antimicrobial activity. The cationic nature of CAMPs facilitates their interaction and movement through the anionic cell membrane of bacteria (Hancock, 2001), and their amphipathic nature enables them to adopt specific conformations that promote their penetration of the bacterial cell and prevents self-aggregation (Yin et al., 2012). Following a primary accumulation of CAMPs at the bacterial cell membrane, various mechanisms have been suggested for explaining their antimicrobial activity, including target of the cell membrane (transmembrane pore model, carpet pore model), cell wall synthesis, intracellular processes, or immune modulation (Kumar et al., 2018). Although the presence of both a hydrophobic core and a net positive charge on the peptide are crucial for this bioactivity, a threshold for both parameters exists above which the activity remains constant, but the hemolytic activity towards mammalian membrane increases (Dathe et al., 2001; Yin et al., 2012). For instance, Yin et al., (2012) observed that modifying the synthetic highly active CAMP 6K-F17 to the more hydrophobic 6K-F17-4L caused a decrease in antimicrobial activity and increase in hemolytic activity. The antimicrobial activity was however restored when positive charges were redistributed on the two termini of the same peptide, generating the peptide 3K-F17-4L-3K. On the other hand, the distribution of charges at the two edges of the native peptide, by generation of the CAMP 3K-F17-3K caused a significant drop in antimicrobial activity. The increased hydrophobic core was likely promoting the self-aggregating β-sheets structures at the bacterial membrane surface, therefore decreasing membrane penetration by the peptide. In a similar study, Dathe et al. (2001) observed a threshold value of +5 in the charge of CAMPs, above which a marked increase in hemolytic activity and decrease in permeabilization of the bacterial membrane was observed. Net charges higher that +5 were possibly causing such a powerful interaction of the peptide with the anionic lipids in the bacterial membrane bilayer that would prevent the peptide transport through the membrane and to reach the target sites. Interestingly, membrane permeability could then be restored by modification of the peptide hydrophobicity. Thus, the design of an optimally active CAMP (i.e., high antimicrobial and low hemolytic activities) is hinged on multiple factors such as peptide helicity, hydrophobicity of the core motifs, distribution of positive charges, reduction in aggregation, and the ability of the peptides to undergo oligomerization in the membrane (Yin et al., 2012).
Because of selectivity of interaction with components of the bacterial cell membrane and low affinity for mammalian membranes, CAMPs have attracted significant interest in recent years as possible substitutes for antibiotics. Also, the non-specific nature of their activity is favorable in preventing bacteria resistance to the peptides (Hancock, 2001). Despite the promising properties, only a few CAMPs have successfully been approved for use by the U.S. Food and Drug Administration, and some others are currently undergoing phase II and III clinical trials. Several challenges still need to be addressed, such as the susceptibility of the peptide to hydrolysis by digestive enzymes, their intrinsic toxicity, and discrepancy between activities in vitro and in vivo. For instance, the most active CAMP in vitro, polyphemusin, was found to be inactive in vivo, whereas some weakly active CAMPs in vitro displayed strong in vivo activities (Hancock, 2001). Currently, the role of peptide hydrophobicity in this instance is not apparent, although emerging work is focusing on using chemical and physical methods to improve the peptide stability by incorporating peptidomimetics and D-amino acids, cyclization, and use of targeted delivery systems (Kumar et al., 2018). Additionally, bioinformatic tools are rapidly evolving for organising large amounts of information in easily accessible databases. Further insights into structure-function relationship of CAMPs and new research directions were recently reviewed by Ahmed and Hammami (2018) and Kumar et al. (2018).
4.3. Bile acid binding capacity
Endogenous cholesterol level is regulated by harmonizing the absorption of food-derived cholesterol and hepatic cholesterol biosynthesis. The absorption of dietary cholesterol is mediated by its solubilization in bile acids, which triggers the formation of micelles that are easily absorbed in the small intestine (Wilson and Rudel, 1994). Bile acid binding peptides have the potential to inhibit cholesterol absorption in the intestinal cells by diminishing bile acid availability and the subsequent incorporation of cholesterol into absorbable micelles. Bile acid binding peptides can also increase bile acid excretion, thereby inhibiting enterohepatic circulation of bile acids and increasing metabolism of hepatic cholesterol to replenish the excreted bile acids (Boachie et al., 2018). Interestingly, Choi et al. (2002) identified a 47-amino acid residue region subunit A1a that appeared to be primarily involved in the bile acid-binding property of soy glycinin. Further analysis led to the identification of an 8-amino acid hydrophobic region (Val-Ala-Trp-Trp-Met-Tyr; also known as soystatin), which is responsible for the bile acid-binding property. Further studies demonstrated that the bile acid-binding by soystatin was translated in vivo in rats (Nagaoka et al., 2010). Based on soystatin, Ito et al. (2018) used in silico analysis, where each amino acid residue was replaced, to demonstrate the additional role of electrostatic properties in bile acid binding. Bile acids are amphipathic and, similar to CAMPs, peptides with hydrophobic and cationic characters are better positioned to respectively interact with the hydrophobic and hydrophilic regions of bile acids. Moreover, considerable attention should be given to the peptide conformation and 3-dimensional structure. For instance, a two-fold increase in bile acid-binding capacity was observed when a chicken meat protein hydrolysate was converted to plastein aggregate, even when the samples have a similar amino acid profiles. This effect is attributed to peptide aggregation and subsequent increase in surface hydrophobicity as a result of plastein aggregate formation (Udenigwe et al., 2015). Furthermore, plastein isolation from the crude reaction product resulted in higher mean particle size, surface hydrophobicity, and hydrophobic-to-hydrophilic amino acid ratio, leading to a higher binding capacity and stronger affinity (lower dissociation constant, Kd) of the aggregated peptides for sodium deoxycholate (Mohan and Udenigwe, 2015). Another hydrophobic peptide, Ile-Ala-Val-Pro-Gly-Glu-Val-Ala, derived from pepsin hydrolysis of 11S globulin was also reported to bind cholic and deoxycholic acids in vitro (Pak et al., 2005), whereas Leu-Pro-Tyr-Pro-Arg (from soybean glycinin) and His-Ile-Arg-Leu (from β-lactoglobulin) decreased serum cholesterol by >25% and 17%, respectively after two days of oral administration to mice (Yoshikawa et al., 2000). Although further research is needed to elucidate in vivo mechanism of bile acid-binding peptides, current evidence indicates the dominance of hydrophobic residues in the peptide structure and supports their important role in the bioactivity.
4.4. Antioxidant capacity
Oxidation is an important process that leads to the generation of free radicals and reactive oxygen species, which are actively involved in physiological processes in the human body. However, excessive oxidation can be damaging to cells and food products. For instance, lipid peroxidation generates unwanted products including aldehydes, peroxides, and ketones (Ayala et al., 2014). Oxidative stress occurs when endogenous antioxidants are overwhelmed by prooxidants, and this can result in disease conditions such as diabetes, arthritis, inflammation, and cancer (Huang et al., 2005). Peptides are known to have strong antioxidant capacity. A study suggested that typical structural characteristics of antioxidant peptides include the presence of hydrophobic amino acid residues Leu or Val at the N-terminus and the presence of Pro, His or Tyr in the sequence (Sabeena Farvin et al. 2010). According to Zou et al. (2016), Trp, Phe, Val, Ile, Gly, Lys, and Pro are the most established hydrophobic amino acids associated with antioxidant activities. Tian et al. (2015) demonstrated that the presence of hydrophobic Trp residues in peptides contributes to high antioxidant capacity. Likewise, Pro residue is thought to alter the secondary structure of peptides, thereby enhancing the interaction of the amino acid residues of peptide and their antioxidative properties (Zou et al., 2016). Additionally, Leu and the motifs Ser-Leu, Thr-Leu and Pro-Leu, especially when located at the N- and C- terminal, are known to enhance the antioxidant activities of peptides, with the C-terminus being more influential in determining the activity (Li and Li 2013). Moreover, hydrophobic Phe and Tyr residues of peptides have aromatic rings capable of reacting with hydroxyl radical to form stable hydroxylated derivatives (Pownall et al., 2010). Apart from this mechanism, hydrophobic residues and motifs are thought to facilitate the interaction of antioxidant peptides with hydrophobic, biological targets of oxidation. A chemometric study reported a positive relationship between the total content of hydrophobic amino acids of food protein hydrolysates and their capacity to scavenge a hydrophobic synthetic radical and hydrogen peroxide (Udenigwe and Aluko, 2011). Despite the experimental evidence to support this relationship, in most cases, the particular role of hydrophobicity in promoting the antioxidant capacity of peptides is still not clear. Table 3 shows a list of some hydrophobic, food protein-derived peptides and their respective in vitro and in vivo bioactivities.
 Click to view | Table 3. Food protein-derived bioactive peptides, with highlighted hydrophobic regions, and their respective bioactivities |
| 5. Conclusion | ▴Top |
Hydrophobicity is an important feature of peptides that determines their interaction with several physiological targets and their bioactivities. Peptide hydrophobicity can be affected by the degree of hydrolysis, processing conditions, type of protease, average hydrophobicity value of protein precursors, and sequence of hydrophobic amino acid residues in the peptide chain. Particular proteases (e.g. Alcalase, thermolysin) have high specificity in cleaving hydrophobic amino acid residues in food proteins during the generation of bioactive peptides, with a concomitant release of hydrophobic peptides and amino acids and increase in the bitterness score. Hydrophobicity is also demonstrated to influence peptide self-assembly, emulsifying capacity, bitterness property and other properties, including biostability and potentially bioavailability. Thus, the ability of bioactive peptides to escape proteolytic degradation and be transported across the intestinal barrier in humans may be enhanced by modifying peptide structural features, including hydrophobicity.
Acknowledgments
We acknowledge the support of the Natural Sciences and Engineering Research Council of Canada (NSERC), reference number RGPIN-2018-06839. Cette recherche a été financée par le Conseil de recherches en sciences naturelles et en génie du Canada (CRSNG), numéro de référence RGPIN- 2018-06839.
Conflict of interest
There is no conflict of interest involved in this article.
| References | ▴Top |