| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Original Research
Volume 4, December 2018, pages 117-122
Facile synthesis of bioactive Allitol from D-psicose by coexpression of ribitol dehydrogenase and formate dehydrogenase in Escherichia Coli
Hinawi A.M. Hassanina, b, Mohammed A.A. Eassaa, b, c, Bo Jianga, b, *
aState Key Laboratory of Food Science and Technology, Jiangnan University, 1800 Lihu Avenue, Wuxi 214122, China
bInternational Joint Laboratory on Safety, Jiangnan University, 1800 Lihu Avenue, Wuxi 214122, China
cDepartment of Food Processing, Faculty of Engineering, University of El Imam El Mahadi, P.O. Box 209, Kosti, Sudan
*Corresponding author: Bo Jiang, State Key Laboratory of Food Science and Technology, Jiangnan University, 1800 Lihu Avenue, Wuxi 214122, China. E-mail:
DOI: 10.31665/JFB.2018.4167
Received: November 4, 2018
Revised received & accepted: December 20, 2018
| Abstract | ▴Top |
Coexpression of formate dehydrogenase (FDH) and ribitol dehydrogenase (RDH) in Escherichia coli was used for the synthesis of Allitol from D-psicose. FDH was coexpressed with RDH for continuous NADH regeneration. The results revealed that the optimum conditions for allitol production occurred at pH 7.0 and 30 °C. Allitol reached the maximum yield of 19.2 mg at 2.0% substrate concentration after 48 hours. Using D-psicose as a substrate, allitol was successfully produced using an engineered E. coli coexpressed with RDH and FDH.
Keywords: Formate dehydrogenase; Ribitol dehydrogenase; Allitol; D-psicose
| 1. Introduction | ▴Top |
Whole-cell biotransformation has many benefits at the commercial manufacturing of rare sugars. For example, the enzyme cofactors (i.e. NAD+ and NADH) can be recycled in vivo and no need to purify the enzymes and thus avoid complications in the process of enzyme purification. In addition, the separation of the final products is easier when using whole-cell fermentation, since the enzyme inside the cell is isolated from the final product by the membrane of the microorganism cell. Crystallization is one of the most important processes in the synthesis of sugars, by using the whole-cell biotransformation the process of crystallization can be performed directly after the end of fermentation. In recent years, many rare sugars have been produced by whole-cell biotransformation using wild-type microorganisms and recombinant strains. This biotransformation have been industrially applied in large quantities, such as the synthesis of D-mannitol using engineered Escherichia coli and C. glutamicum ATCC13032 (Kaup et al., 2004; Wu et al., 2013; Zhao et al., 2013; Zhou et al., 2012).
Alitol is a rare sugar of great importance due to the properties it possesses, such as anti-obesity, and laxative effects for the treatment of constipation, which the water content of the lumen is increased significantly by allitol. In addition, allitol cross links between l/d-hexoses in Izumoring strategy (Hassanin et al., 2017). It was chemically synthesized using sodium borohydride as an oxidizing agent and D-psicose as substrate (Ballard and Stacey, 1973). However, the chemical methods used for rare sugars production are not industrially effective due to the presence of impurities in the final product.(Ballard and Stacey, 1973; Takeshita et al., 2000; Takeshita et al., 1996). According to the allitol position as a bridge between D-psicose and L-psicose and its chemical structure as alcoholic sugar, d-psicose can be reduced into allitol by using ribitol dehydrogenase (RDH), this reaction required NADH as a cofactor. NADH is an expensive cofactor, which can limit the use of enzymatic reaction for allitol production. Formate dehydrogenase (FDH) is an enzyme that responsible for regeneration of NADH from NAD+ using sodium formate as substrate. FDH and RDH were coupled for allitol production to overcome the NADH regeneration problem. Several studies have used whole-cell biotransformation for production of allitol, in an attempt to overcome the cofactor regeneration problem. Such as Enterobacter agglomerans strain 221e was used for allitol synthesis from D-psicose (Muniruzzaman et al., 1995).
Expression systems in mammalian cell culture, yeast and baculovirus are very expensive. However, the coexpression system in E. coli is commonly applied to overcome difficulties of heterologous protein expression (Tolia and Joshua-Tor, 2006). The coexpression in E. coli provides a useful alternative system for production of rare sugars. Zhu et al. (2015) reported production of allitol from D-fructose using an engineered E. coli strain expressing membrane protein glucose/fructose facilitator (GLF), D-psicose 3-epimerase (DPE), FDH, and RDH. The GLF was used to increase the intracellular D-fructose concentration (Parker et al., 1995; Weisser et al., 1995).
FDH and RDH enzymes isolated from Ogataea parapolymorpha DL-1 and Providencia alcalifaciens RIMD 1656011, respectively, were previously cloned, characterized, and coupled for production of allitol from D-psicose (Hassanin et al., 2016a; Yu et al., 2014). In this present work, we studied the production of allitol from d-psicose using whole-cell biotransformation carried out by engineered Escherichia coli co-expressed with RDH and FDH from O. parapolymorpha DL-1 and P. alcalifaciens RIMD 1656011, respectively. All the optimal conditions for the production were optimized to produce the allitol at low cost.
| 2. Materials and methods | ▴Top |
2.1. Equipment
The entire experimental process required, FE20 pH meter, Vibra-Cell™ 72405 sonicator (BioBlock Scientific, Illkirch, France), DHG 9070A oven (Shanghai, China), Mettler Toledo PL 2002 balance (Shanghai, China), Hook biological safety cabinet, HYG-2—incubator for liquid culture (Shanghai, Chaina), DK-S24—water bath, Eppendrof 5804 R—centrifuge (Hamburg, Germany), Alpha-1 spectrophotometer, NANODROP 2000C—nanodrop (USA), CL-40M Autoclaves (Steam sterilizer), and Agilent 1200 series—equipment for HPLC (Shodex, Tokyo, Japan).
2.2. Strains and coexpression of RDH and FDH
The host strain used for gene cloning was E. coli DH5α while for coexpression was E. coli BL21. The pETDuet-1 was used as the coexpression vector. The FDH and RDH genes were from Ogataea parapolymorpha DL-1 and Providencia alcalifaciens RIMD 1656011, respectively (Table 1) (Hassanin et al., 2016a; Yu et al., 2014). Luria-Bertani medium (LB) was used for growth of all strains. All strains were cultivated at 37 °C and induced by 1.0 mmol L−1 isopropyl β-d-1-thiogalactopyranoside (IPTG) present in the medium at 20 °C.
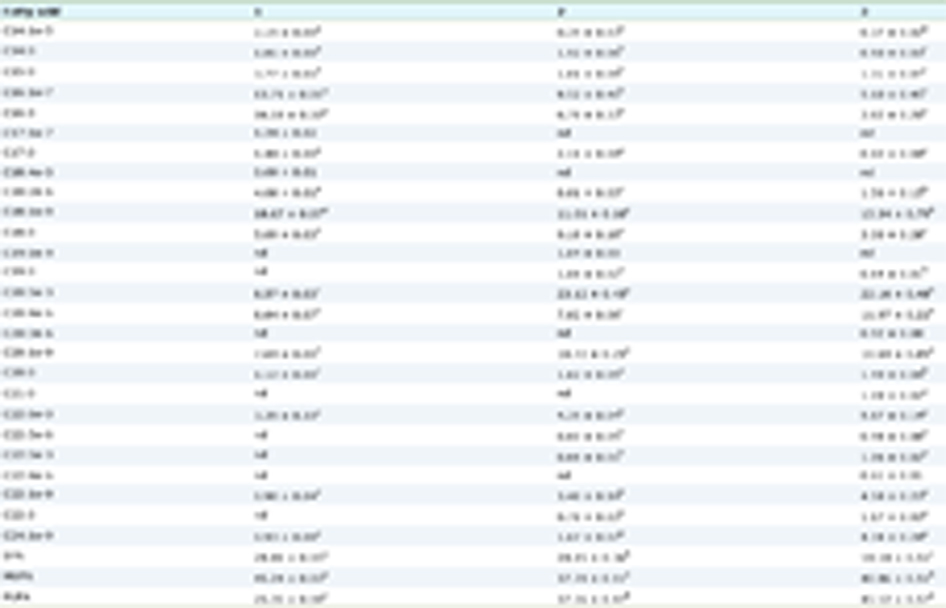 Click to view | Table 1. Plasmids and strains used in this study |
FDH and RDH genes were synthesized by Sangon Biological Engineering Technology and Services (Shanghai, China). They were multiple cloned into pETDuet-1 using Generay Biotechnology (Shanghai, China) to create pETDuet-RDH-FDH (figure 1). The RDH gene was amplified and ligated into the first multiple cloning sites of pETDuet-1 at the SalI and NotI site to create the recombinant plasmid, named pETDuet-RDH. A 6 × His-tag sequence was present in the vector to aid protein purification. The FDH gene was amplified and ligated into the second site of pETDuet-RDH at the NdeI and XhoI site to create the recombinant plasmid pETDuet-RDH-FDH.
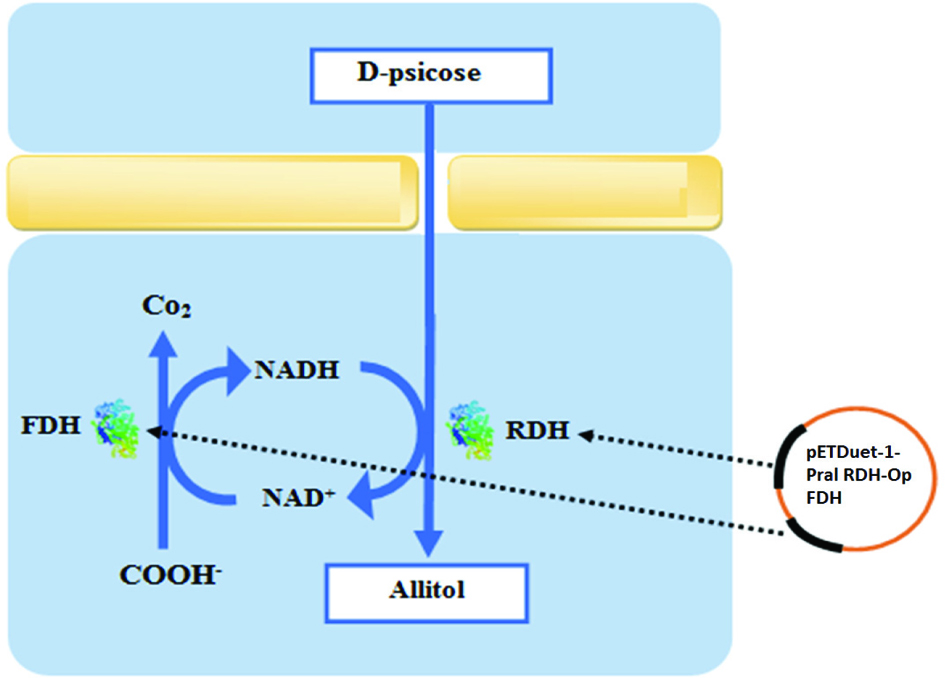 Click for large image | Figure 1. Allitol production pathway using engineered E. coli coexpressed with ribitol dehydrogenase (RDH) and formate dehydrogenase (FDH). |
The pETDuet-RDH-FDH plasmid was expressed into E. coli BL21(DE3) to form an engineered strain BL21 Star (DE3) pETDuet-RDH-FDH, which grown in LB medium supplemented with ampicillin (100 μg mL−1). When the cells growth reaches an optical density (600 nm) of 0.6 at 37 °C, IPTG was added to the culture to a final concentration of 1 mmol L−1, then FDH and RDH was induced at 20 °C for 20 h.
The subunit molecular mass of RDH and FDH extracted form engineered E. Coli were examined using the denaturing conditions of sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) on 50 g L−1 stacking gel and 120 g L−1 separating gel.
2.3. Optimization of temperature, pH and cell concentration
Whole-cell biotransformation was carried out with 1% D-psicose, 1% sodium formate, 2 mmol L−1 NAD+, three buffer systems, i.e. phosphate buffer (50 mmol L−1, pH 5.0–0.7), Tris-HCl (50 mmol L−1, pH 7.0–9.0), and glycine-NaOH (50 mmol L−1, pH 0.9–11.0), were used to determine the optimal pH for the whole-cell biotransformation. The optimal temperature was measured over the temperature range of 25–45 °C at intervals of 5 °C. To detect the optimum biomass, different cell concentrations (OD600 = 20, 40, 60, and 80) were used in the biotransformation.
2.4. Allitol production using whole-cell biotransformation
The engineered E. coli strain was centrifuged to collect the precipitate cells and washed twice with 50 mmol L−1 Tris-HCl buffer (pH 7.0). The washed cells were suspended in 50 mmol L−1 Tris-HCl buffer (pH 7.0) containing 0.5, 1, and 2%, D-psicose. The biotransformation was conducted at 30 °C and 200 rpm. Samples were collected at different times to measure the concentration of allitol produced.
2.5. Quantification of D-psicose and allitol
D-psicose and allitol concentrations in the biotransformation mixtures were examined by high-performance liquid chromatography (HPLC). The mixtures were filtered through 0.22 μm membrane filters. SUGAR-PAK column (6.5 × 300 mm) was used for separation of the products. the column conditions temperature and flow rate were 85 °C and 0.4 mL min−1 using water as mobile phase.
| 3. Results and discussion | ▴Top |
The responsible enzyme for production of allitol from d-psicose is RDH enzyme. In a previous study, P. alcalifaciens RDH was cloned, characterized and the crude enzyme was used for allitol production from D-psicose (Hassanin et al., 2016b). The SDS-PAGE result analyses for RDH from P. alcalifaciens showed that the subunit molecular mass around 25 kDa. FDH the enzyme that responsible for the regeneration of NADH was cloned and characterized by Yu et al. (2013). The FDH subunit molecular mass was around 40 kDa. In a previous study, crude RDH and FDH were coupled for the synthesis of allitol from d-psicose, which the optimum temperature and pH were 40 °C and pH 7.5, respectively (Hassanin et al. 2016b).
In this study, FDH and RDH genes were sub-cloned into two sites of pETDuet-1 (figure 2a) and coexpressed in E. Coli. A reaction was conducted with the extracted DNA from engineered E. coli to confirm successfully sub-cloned vector (pETDuet-1) with RDH and FDH genes, as shown in figure 2b the actual electrophoreses gel of target DNA extracts and digested with restriction enzymes (EcoRI/XhoI) confirmed the successfully cloning steps.
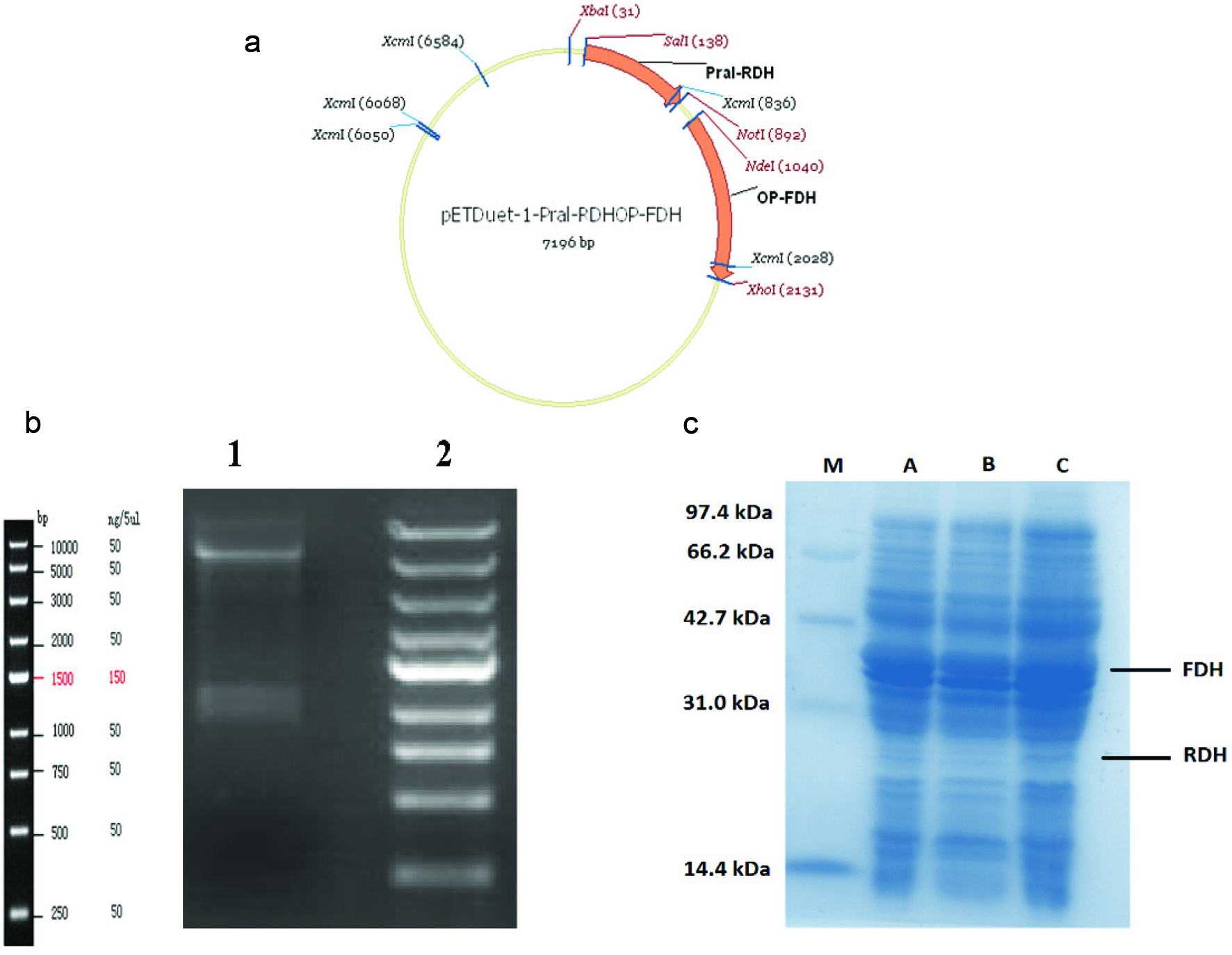 Click for large image | Figure 2. Plasmid construction (a) constructed vector map (pETDuet-RDH-FDH). (b)The result of extracts DNA from transformed E. coli with pETDuet-RDH-FDH (1) DNA ladder (2) The DNA digested with EcoRI/XhoI restriction enzymes. (c) SDS-PAGE analysis of the co-expression recombinant enzymes. (M) protein marker; (A) proteins in the supernatant of constructed E. coli induced with IPTG (6 hours), (B) proteins in the supernatant of constructed E. coli without IPTG; (C) proteins in the supernatant of constructed E. coli induced with IPTG (20 hours). |
To produce genetically engineered E. coli for the synthesis of allitol from d-psicose, pETDuet-RDH-opFDH contained RDH and FDH genes was coexpressed into E. coli. RDH and FDH genes were combined to form production and cofactor recycling pathway. The engineered E. coli coexpressed with recombinant plasmid pETDuet-RDH-FDH was induced by IPTG. As shown in figure 2c SDS-PAGE results for the cell lysate of the engineered E. coli revealed two bands corresponding to RDH, and FDH proteins.
To increase the productivity of specific substrate such as allitol in whole cell biotransformation, supplementation with NADH is needed (Kataoka et al., 1999; Kizaki et al., 2001; Yamamoto et al., 2003). Since NADH is very expensive, this increases the cost of production. However, the combination of FDH with RDH to construct cofactor recycling system for regeneration of NADH from NAD+ can ensure the continuous supply required for d-psicose reduction. The cofactor recycling system required supplementation with sodium format, which is inexpensive. Therefore, it significantly reduces the cost of allitol production.
Conditions of biotransformation using engineered E. coli were optimized to achieve the high yield of allitol. The effect of temperature was examined on the biotransformation at different temperatures (25, 30, 35, 40 and 45 °C), and the final product allitol was determined by using HPLC. As shown in figure 3a when the temperature increased, the yield of allitol increased until the temperature reached 30 °C and then decreased. As previously characterized the optimum temperatures for RDH and FDH were 35 and 65 °C, respectively (Hassanin et al., 2016a; Yu et al., 2014). However, the optimal temperature for whole-cell biotransformation was registered at 30 °C, which is near to the optimum temperature for RDH. Han et al. (2014) reported production of allitol from D-psicose using Klebsiella oxytoca G4A4 isolated from soil and the optimal temperature was 37 °C. Similarly, Zhu et al. (2015) reported production of allitol from D-fructose using an E. coli strain expressing D-psicose 3-epimerase (DPE), FDH and RDH and the optimum temperature was 40 °C.
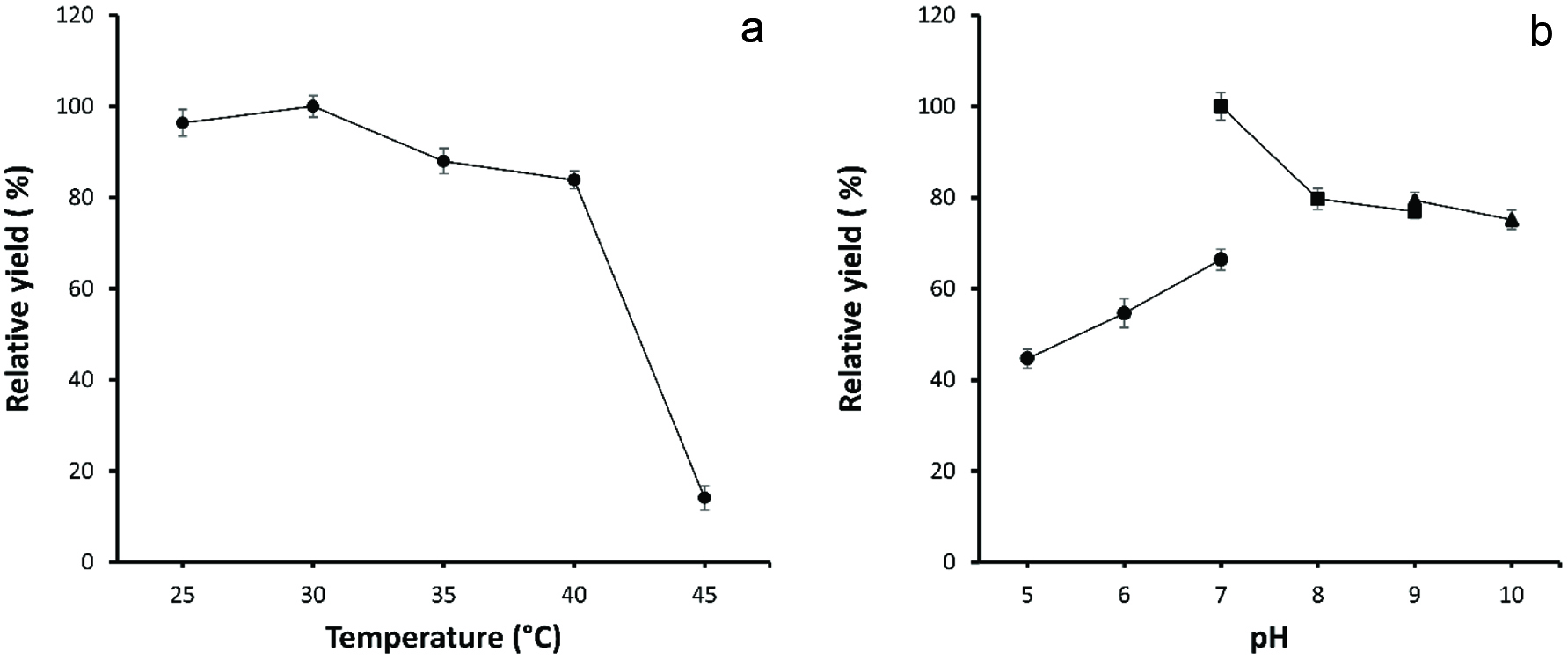 Click for large image | Figure 3. Effect of temperature (a) and pH (b) ((•) Phosphate buffers (■) Tris-HCl buffers (▲) Glycine-NaOH buffers) on the whole cell biotransformation. |
The biotransformation was conducted at various pH values, to study the effect of pH on allitol synthesis. As shown in figure 3b, the optimum pH for allitol production was registered at pH 7.0 in 50 mmol L−1 Tris-HCl buffer. This low optimum pH value may due to reducing of formic acid, which resulting in gradually increasing in pH value during biotransformation (Kaup et al., 2004; Zhu et al., 2015). However, the optimum pH for synthesis of allitol using Klebsiella oxytoca G4A4 was 8.0, while the optimum pH of the engineered E. coli with (DPE), FDH and RDH was 7.0 (Han et al., 2014; Zhu et al., 2015).
To optimize the cell biomass, the resting cells at different biomasses (OD600 = 20, 40, 60, and 80) were previously prepared and used for the synthesis of allitol from D-psicose. Figure 4 showed that the allitol yield increased from OD600 = 20 to OD600 = 80, as results show that the rate of increase of the allitol yield did not increase linearly. When the cell biomass reached OD600 = 40, the rate of increase dropped. We selected a cell biomass of OD600 = 40 as the optimum cell concentration for allitol synthesis to reduce the production cost.
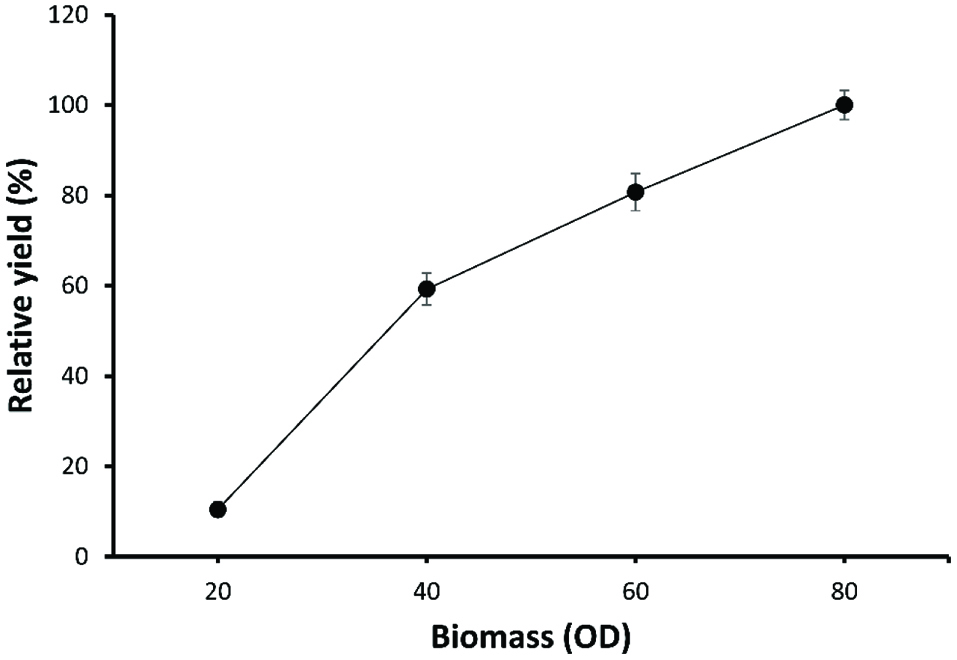 Click for large image | Figure 4. Effect of the cell biomass on the synthesis of allitol using whole cell biotransformation. |
Production of allitol was conducted at different concentrations of d-psicose (0.5, 1.0 and 2.0%). As shown in figure 5 the reduction rates of d-psicose were about 7.3, 10.0 and 19.2 mg at 0.5, 1.0 and 2.0%, respectively after 48 hours. At a concentration of 0.5 and 1.0% D-psicose, maximum reduction was registered after 48 hours of biotransformation, while at 2.0% D-psicose concentration, maximum reduction was found at 72 hours. However, after12 h the reduction rates were 2.9, 4.3 and 7.8 mg for 0.5, 1.0 and 2.0% D-psicose, respectively. Reduction rates were measured from the concentration of allitol produced in the biotransformation mixture.
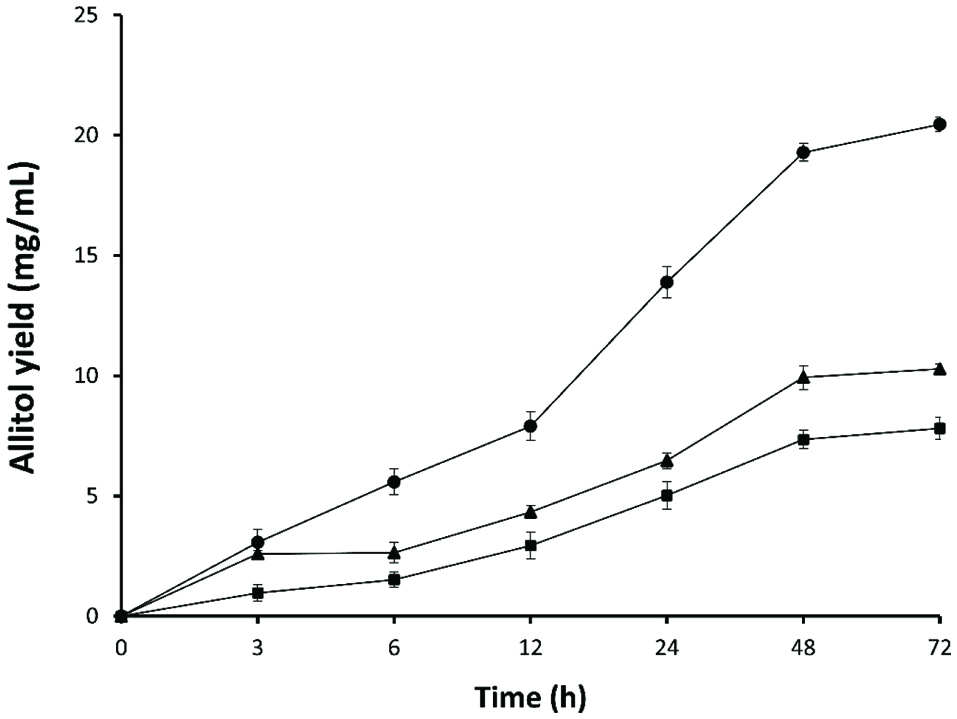 Click for large image | Figure 5. Effect of cell biomass on allitol synthesis. (■) 0.5% D-psicose. (▲) 1% D-psicose (•). 2% D-psicose. |
The use of whole-cell biotransformation instead of enzyme reaction system for the production of allitol has several advantages. As we mentioned the RDH is an enzyme that responsible for the synthesis of allitol from d-psicose, and this reaction required NADH as cofactor, which is very expensive. FDH was coupled with RDH for continuous regeneration of NADH using sodium formate as substrate. There are several disadvantages in using the enzyme reaction. For example, enzyme extraction and separation of the final product needs several steps. Whole-cell biotransformation of engineered E. Coli overcome the disadvantages of the enzyme reaction system.
| 4. Conclusions | ▴Top |
In this study, an engineered E. coli for the synthesis of allitol from D-psiocse was constructed. The multi-enzyme biotransformation pathway was successfully used for the production of allitol by coexpression of RDH and FDH in E. Coli. FDH was used to regenerate NADH from NAD+ inside the engineered E. Coli to decrease the cost of allitol production. the maximum yield of allitol was 96 % using the whole-cell biotransformation system. Based on these results, this engineered E. coli enormous potential for application in the production of allitol.
Conflict of interest
The authors declare no conflict of interest.
This work was supported by National Key Research and Development Program (2017YFC1600902) and National Natural Science Foundation of China (31871745).
| References | ▴Top |