| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Original Research
Volume 1, March 2018, pages 153-165
Protein extraction and bioactive hydrolysate generation from two microalgae, Porphyridium purpureum and Phaeodactylum tricornutum
Julianne Stacka, Aurélien V. Le Gouica, Paul R. Tobina, Freddy Guihéneufb, Dagmar B. Stengelb, Richard J. FitzGeralda, *
aProteins and Peptides Research Group, Department of Biological Sciences, University of Limerick, Ireland
bBotany and Plant Science, School of Natural Sciences and Ryan Institute for Environmental, Marine and Energy Research, National University of Ireland Galway, Galway, Ireland
*Corresponding author: Richard J. FitzGerald, Proteins and Peptides Research Group, Department of Biological Sciences, University of Limerick, Ireland
DOI: 10.31665/JFB.2018.1134
Received: January 26, 2018
Revised received & accepted: March 2, 2018
| Abstract | ▴Top |
Microalgae are a relatively underutilised source of valuable nutritional compounds and biochemicals. Their high protein contents make them an interesting target for the generation of bioactive peptides for the functional food industry. Here, the nitrogenous components of the red microalga Porphyridium purpureum and the diatomPhaeodactylum tricornutum were quantified, and their SDS-PAGE profiles analysed. Proteinswere isolated from these species by solubilisation at alkaline pH followed by precipitation at acidic pH. The protein extracts were hydrolysed with the food-grade proteolytic preparations Alcalase 2.4 L and Flavourzyme 500 L. The P. purpureum and P. tricornutum hydrolysates were examined for their in vitroanti-diabetic and antioxidant properties. The microalgal-derived protein hydrolysates had dipeptidyl peptidase (DPP) IV inhibitory activity, with IC50 values of 2.28 ± 0.36 and 2.68 ± 0.19 mg/mL for P. purpureum and P. tricornutum, respectively. The oxygen radical absorbance capacity (ORAC) and ferric reducing power (FRAP) values were, respectively, 13.98 ± 0.97 and 478.94 ± 34.43 for P. purpureum, and 15.04 ± 0.54 and 155.74 ± 38.30 for P. tricornutum expressed in µmol Trolox Equivalent (TE) per gram of dry matter.The results suggest that protein hydrolysates from these microalgae have potential use as functional food ingredients.
Keywords: Porphyridium purpureum; Phaeodactylum tricornutum; bioactive peptides; protein hydrolysates; antioxidant; DPP-IV
| 1. Introduction | ▴Top |
Of the over 20,000 different species of microalgae, only a small number have been characterized (Guiry, 2012). Microalgae are an underutilised resource of valuable bioactive compounds and biochemicals such as proteins, pigments, antioxidants, polysaccharides, sterols, fatty acids and vitamins. Microalgae contribute indirectly to the human diet due to their frequent use as feed additives in aquaculture and agriculture(Hayes et al., 2017). Additionally, some microalgae such as Aphanizomenon flos-aquae, Chlorella vulgaris and Spirulina platensis are consumed directly in the human diet as whole-biomass because of their good nutritional properties (Saha et al., 2015). The high protein content of various microalgal species and their amino acid profile, which compares favourably with that of other food proteins, make microalgae an alternative human food protein source (Hayes et al., 2017). Spirulina, for instance, is high in protein (60–70% depending on the strain). Not only does this protein possess all of the essential amino acids, but these amino acids are reported to have excellent bioavailability (Lourenco et al., 2004; Plaza et al., 2009).
Studies on microalgal proteins, to date, have mainly focused on evaluating the nitrogen to protein (NTP) conversion factor (Lourenco et al., 1998; Lourenco et al., 2004; Safi et al., 2013); finding the best methods of protein extraction and quantification (Barbarino and Lourenço, 2005; Safi et al., 2013; Safi et al., 2014); and analysis of the behavior of proteins at the air/water interface (Chronakis et al., 2000). However, proteins from marine sources show much promise as functional ingredients in foods because they possess numerous important and unique properties such as good foaming capacity, gel forming ability and antimicrobial activity (Rasmussen and Morrissey, 2007; Ovando et al., 2018). In addition, many food proteins contain, encrypted within their primary structures, peptide sequences capable of modulating specific physiological functions referred to as bioactive peptides. These peptides are liberated by the action of proteinases including pepsin, Alcalase, Flavourzyme, and papain among others, and can have antioxidant, anti-hypertensive, and/or anti-diabetic properties among others (Nongonierma et al., 2016). Microalgae have already been shown to be a source of bioactive peptides. Peptides purified from Chlorella vulgaris have demonstrated significant antioxidant effects(Sheih et al., 2009b), angiotensin I-converting enzyme (ACE) inhibitory activity(Suetsuna and Chen, 2001) and anti-proliferative activity (Sheih et al., 2009a; Wang et al., 2010).An Alcalase hydrolysate fromChlorella ellipsoidea yielded a peptide (VEGY) with potent ACE inhibitory activity and oral administration of this peptide significantly decreased systolic blood pressure in spontaneously hypertensive rats (Ko et al., 2012). The tripeptide TRY isolated from a thermolysin hydrolysate of Palmaria palmata, had an ACE half maximum inhibitory concentration (IC50) value of 0.044 μmol (Furuta et al., 2016).The tetrapeptide, EDKR, from the diatom Navicula incerta, was shown to possess antioxidant activity(Kang et al., 2011), while other peptides from this organism displayed anti-hepatotoxic effects (Kang et al., 2013). Furthermore,anti-inflammatory peptides have been purified from enzymatic hydrolysate of Spirulina maxima (Vo et al., 2013), andS. platensis peptides have been reported to possess anti-hypertensive, antiallergic, anti-inflammatory, anti-atherosclerotic, anti-tumor and antiaging activities (Ovando et al., 2018).
Bioactive peptides in food have the potential to provide health benefits beyond the basic nutritional value of the product, and may find use in the prevention and management of a wide range of diseases. Type 2 diabetes mellitus (T2DM) is a metabolic disorder characterized by both insulin deficiency and peripheral insulin resistance, which causes significant morbidity and mortality and is a considerable burden on health-care resources. The incretin hormones, glucagon-like peptide 1 (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP), are rapidly cleaved and rendered metabolically inactive by the action of dipeptidyl peptidase-IV (DPP-IV, Deacon et al., (1995)). Inhibition of this enzyme has therefore become a new approach for the treatment of T2DM. A number of food protein-derived peptides with DPP IV inhibitory activity have been described (Van Amerongen et al., 2009; Harnedy and FitzGerald, 2013b; Lacroix and Li-Chan, 2013; Nongonierma and FitzGerald, 2017), but to date there appears to be no information regarding the ability of microalgal-derived protein hydrolysates to inhibit DPP IV activity. Oxidative stress plays a key role in T2DM complications including atherosclerosis, retinopathy and nephropathy (Rochette et al., 2014; Oseguera-Toledo et al., 2015). Antioxidants protect internal organs and tissues from oxidative damage by toxic reactive oxygen and nitrogen species (Ahn et al., 2004), and the market demand for natural antioxidants is increasing (Saha et al., 2015).
Thered microalgal genus Porphyridium is of commercial interest as a source of bioactive compounds such as phycobiliproteins, sulphated exopolysaccharides and long-chain polyunsaturated fatty acids (LC-PUFAs) (Spolaore et al., 2006). Phycobiliproteins are used as dyes for food and cosmetics (Spolaore et al., 2006), and also possess several therapeutically interesting bioactivities, namely anticancer, antioxidant, hepato-protective and immunomodulating effects (Plaza et al., 2009). To our knowledge there are no reports, to date, on the bioactivity of protein hydrolysates from Porphyridium spp. The biomass of diatoms such as P. tricornutumis reported to contain substantial levels of protein (35–66% d.w. Fernández-Reiriz et al., (1989)), and they are currently being investigated as expression systems for various recombinant proteins (Hempel et al., 2011). Similar to P. purpureum, thus far there appears to be no reports on the bioactivity of protein hydrolysates from P. tricornutum.
The aim of this investigation was to examine the in vitro antioxidant and DPP-IV inhibitory abilities of protein hydrolysates from the commercially relevant microalgal species, P. purpureumand P. tricornutum. This studyreports onquantification of the nitrogenous components of these species, as well as on their SDS-PAGE profiles. We also determined the optimal alkaline-soluble protein extraction conditions for P. purpureum and P. tricornutum, and describe the use of proteolytic enzyme preparations to hydrolyse these protein extracts. The bioactivity of these hydrolysates was then determined.
| 2. Materials and methods | ▴Top |
2.1. Microalgal strains and reagents
The red microalga Porphyridium purpureum (PLY#539; taxonomically synonymous to P. cruentumat the National Centre for Marine Algae and Microbiota (NCMA https://ncma.bigelow.org/)was obtained from the Plymouth Culture Collection of Marine Microalgae at the Marine Biological Association of the United Kingdom. Phaeodactylum tricornutum (UTEX#646) was obtained from the Culture Collection of Algae at The University of Texas at Austin (USA). Corolase® PP was provided by AB Enzymes (Darmstadt, Germany), and Alcalase® 2.4 L andFlavourzyme® 500 L were obtained from Novozymes A/S (Bagsvaerd, Denmark). ProtoGel®, ProtoGel® resolving buffer (4x), ProtoGel® stacking buffer (4x), Tris-Glycine-SDSPAGE buffer (10x) and protein loading buffer blue (2x)were from National Diagnostics (Wilmington,USA). Trinitrobenzenesulphonic acid (TNBS) reagent was from Fisher Scientific (Dublin, Ireland). Phenol solution (equilibrated with 10 mM Tris HCl, pH 8.0, 1 mM EDTA) and all other reagents were supplied by Sigma (Arklow, Ireland).
2.2. Growth conditions
Microalgal strains were batch-cultivated in a 70 L flat panel photobioreactor (FP-PBR) using a modified F/2 mediumat 15 °C and under continuous illumination (100 μmol photons m-2 s-1) at the National University of Ireland, Galway. Light was provided on one side of the FP-PBR and cultures were agitated by air-bubbling. F/2 medium used for P. tricornutum culture was composed and complemented as described by Guihéneufand Stengel (2013) with the minor modifications of using natural filtered seawater, an initial NaNO3 concentration of 200 mg/L and no NaHCO3 supplementation. The F/2 medium used for P. purpureumculture also contained a higher initial NaNO3 concentration of 1 g/L. Microalgal cells were harvested gently by centrifugation at 4 °C (1200 × g for 5 min) using a modified Alistar milk cream separator operating at 80L/h. The fresh algal pastes collected were frozen at -20 °C and freeze-dried using a Labconco Freezone® 6 (Kansas City, MO, USA) freeze-dryer system.
2.3. Determination of total Nitrogen (TN), non-protein Nitrogen (NPN) and protein Nitrogen (PN)
The TN content of the microalgal samples was quantified using a modification of the macro-Kjeldahl procedure described by Connolly et al. (2013). Sample (200 mg in triplicate) was weighed out in N free paper (Whatman, B-2 grade) and digested in 20 mL concentrated sulphuric acid with one Kjeldahl catalyst tablet. Initial digestion was for 30 min at 210 °C followed by a further 3 h digestion at 420 °C. After cooling, samples were analysed using the Auto-Kjeldahl System K-370 (BUCHI Labortechnik AG, Flawil, Switzerland). Sodium caseinate of known protein content was used as standard.
Freeze-dried, milled microalgae(∼800 mg in triplicate) were rehydrated in de-ionized water (dH20; 1:20 (w:v)). Samples were stirred overnight at 4 °C to extract the water-soluble components. Samples wereadjusted to 12% trichloroacetic acid (TCA) by treatment with 72% TCA, stirring for 3 h at 4 °C, to precipitate the protein components. Samples were centrifuged at 4,190 × g for 15 min at 4 °C (Hettich Universal 320R, Andreas Hettich GmbH & Co. KG, Tuttlingen, Germany). The pellet and supernatant were separated for Kjeldahl analysis of the PN and NPN fractions as described above.
2.4. Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE)
Whole microalgae were prepared for SDS-PAGE analysis using a method modified by Wang et al. (2006). Dried samples were ground into a fine powder under liquid nitrogen and 0.1 g was transferred into a 1.5 mL Eppendorf tube. Acetone containing 10% TCA (1.5 mL) was added to each tube. After vortexing thoroughly for 30 s, the tubes were centrifuged at 21,250 × g (Hettich Universal 320R) for 3 min at 4 °C. Supernatant was removed. The pellets were resuspended with 1.5 mL 80% methanol containing 0.1 M ammonium acetate, then vortexed and centrifuged as described. The supernatants were removed. The pellets were resuspended with 1.5 mL 80% acetone, then vortexed and centrifuged as described. The supernatants were removed. The pellets were air-dried to remove residual acetone, then re-suspended in 0.5 mL phenol(Tris-buffered,pH8.0) and 0.5 mL SDS buffer(30% (w/v) sucrose, 2% (w/v) SDS, 0.1 M Tris-HCl, pH 8.0, 5% (v/v) 2-mercaptoethanol). The pellets were then vortexed thoroughly for 30 s, incubated on ice for 5 min, and centrifugedat 21,250 × g, for 5 min at 4 °C. The upper phenol phase was transferred to a new 1.5 mL Eppendorf without disturbing the interphase. OnemL100% methanol containing 0.1 M ammonium acetate was added to each tube. The tubes were vortexed and stored at -20 °C for 30 min. The tubes were centrifuged at 21,250 × g for 5 min at 4 °C. The supernatant was removed. Methanol (1.5 mL 100%) was added to each tube. The tubes were vortexed thoroughly for 30 s and then centrifuged at 21,250 × g for 3 min at 4 °C. The supernatants were removed and1.5 mL 80% acetone was added to each tube. The tubes were then vortexed and centrifuged as described. The supernatants were removed. The pellet was air-dried to remove residual acetone and was then re-suspended in Laemmli sample buffer. Protein concentration was measured using the Bradford assay (Bradford, 1976) and 2.5 to 15.0 μg protein was loaded per lane of SDS-PAGE gel using protein loading buffer (National Diagnostics, Wilmington). SDS-PAGE analysis was carried out using a Mini Protean II electrophoresis system (Bio-Rad, Hercules, CA, USA) as described by Harnedy and FitzGerald (2013a).
2.5. Optimisation of protein extraction conditions
The procedures used for the extraction of soluble proteins from milled freeze-dried microalgae were based on a combination of previously published methods (Fleurence et al., 1995; Safi et al., 2014)with some modifications.In brief, freeze-dried milled microalgae was suspended in dH20 (1:20 (w/v)) and gently stirred for 16 h at 4 °C. The protein partitioning to the aqueous phase was removed following centrifugation at 4,190 × g for 15 min at 4 °C. The protein present in the supernatant was termed the aqueous protein extract. The pellet from the above was re-suspended in dH20 (1:50 (w/v)) and the pH was adjusted with 2 M NaOH to pH 9, 10, 11 and 12. The suspension was gently stirred at temperatures ranging from 20 to 50 °C for periods ranging from 1 to 5 h. The effect of pH, duration of extraction and temperature on the total amount of protein recovered was determined. The extract containing alkaline soluble proteins was removed following centrifugation at 4,190 × g for 15 min at room temperature (22 °C). This material was termed thealkaline protein extract. All extractions were performed as independent triplicates (n = 3) from the same biomass and the results were expressed as mg of protein extracted per g dry weight (mean ± SD). SDS-PAGE analysis of aqueous and alkaline protein extracts was performed as described above.
2.6. Protein determination
Protein was determined by a modification of the Lowry protein method as previously described (Harnedy and FitzGerald, 2013a). All samples were analysed in triplicate (n = 3).
2.7. Determination of endoproteinase (EP) activity
EP activity was tested using a modification of the azocasein method (Kilcawley et al., 2002). Corolase PP was used as a positive proteolytic control in all experiments. The reaction mixture containing 700 μL of 0.5 g/100 mL azocasein in 0.05 M phosphate buffer and either 100 μLCorolase PP or 400 μL (microalgal extract) was incubated for 30 min (Corolase PP) or 180 min (microalgal extract). The reaction was terminated by the addition of 100 μL 2 M TCA, mixed and centrifuged at 21,250 × g for 5 min. The resultant supernatant (750 μL) was then mixed with 250 μL 0.5 M NaOH and the absorbance quantified at 440 nm. Each assay was performed in triplicate (n = 3). EP activity was expressed as the change in absorbance (λ = 400 nm)/min/mg of protein.
2.8. Protein enrichment
The microalgal proteins obtained from the aqueous and alkaline extraction procedures were isoelectric precipitated by adjustment with 1 MHCl to pH 2.5 (P. purpureum) or 3.5 (P. tricornutum). The protein isolate was collected after centrifugation at 4,190 × g for 15 min at 20 °C, the pellet being re-suspended in dH20 (0.5 × the original volume of protein extract) and the pH neutralized with 0.1 M NaOH. The isoelectric precipitation and centrifugation process was repeated, after which the pellet was resuspended in dH20 to a protein concentration of 1.6% (w/v) at pH 7.
2.9. Enzymatic hydrolysis of microalgal proteins
Aqueous solutions of the microalgal proteins (1.6% (w/v))were preheatedto 50 °C and adjusted to pH 7 with 0.5 M NaOH. Alcalase and Flavourzyme were each added at anenzyme/substrate (E/S) ratio of 1% (v/w). During hydrolysis (4 h) at 50 °C, the reaction mixture was maintained at pH 7 using a pH-Stat (842 Titrando, Metrohm, Herisau, Switzerland). Heating at 90 °C for 20 min inactivated the proteolytic enzymes. Control protein samples, without added proteolytic preparation, were treated in the same manner. Hydrolysate samples were subsequently freeze-dried.
2.10. Monitoring the extent of hydrolysis and GP-HPLC
The extent of hydrolysis was estimated by the trinitrobenzenesulphonic acid (TNBS) method as described by Harnedy et al. (2013b).SDS-PAGE analysis of protein hydrolysates was performed as described above. The molecular mass distribution of the proteins and peptides was assessed using gel permeation high performance liquid chromatography (GP-HPLC) performed as described by Spellman et al. (2005). The system was calibrated using the following standard proteins: bovine serum albumin (66 kDa), β-lactoglobulin (36 kDa), α-lactalbumin (14.2 kDa), aprotinin (6.5 kDa), bacitracin (1.4 kDa), LWMR (0.605 kDa), DE (0.262 kDa) and tyrosine (0.181 kDa).
2.11. Preparation of samples for bioassays
Freeze-dried protein hydrolysate samples were resuspended in the respective assay buffers at 30 mg/mL. The resultant solutions were diluted prior to bioassay. Due to limited availability of the extracted protein, the analyses were performed in triplicate (n = 3) on a single hydrolysate.
2.12. Bioactivity screening
2.12.1. Dipeptidyl peptidase (DPP) IV inhibitory activity
DPP IV inhibitory activity was determined by measuring free 7-amino-4-methyl-coumarin(AMC) liberated from the fluorogenic substrate Gly-Pro-AMC using the method described by Harnedy et al. (2015). The protein hydrolysate samples were assayed at concentrations between 0.005 and 10 mg/mL. IC50 values (inhibitor concentration that inhibits DPP IV activity by 50%) for each sample were determined by plotting DPP IV inhibition as a function of hydrolysate concentration. The logarithmic regression equation generated from this plot was then used to calculate the IC50 value. The values were expressed as the mean IC50 ± standard deviation (n = 3).
2.12.2. Ferric reducing antioxidant power (FRAP) activity
The FRAP activity of hydrolysate samples was determined using a plate reader as described by Harnedy and FitzGerald (2013b). The FRAP activity of each hydrolysate was expressed as μmol of Trolox equivalents (TE) per gram of freeze-dried powder (FDP) (μmol of TE/gFDP). All assays were performed in triplicate (n = 3).
2.12.3. Oxygen radical absorbance capacity (ORAC) assay
The ORAC assay was performed using a plate reader as described by Harnedy and FitzGerald (2013b). The ORAC value was calculated and expressed as μmol of Trolox equivalents per gram of freeze-dried powder (μmol of TE/g FDP). All assays were performed in triplicate (n = 3).
2.13. Statistical analysis
Statistical analysis was performed using the GraphPad Prism 5 Program (GraphPad, San Diego, CA). The results were analysed by one-way analysis of variance (ANOVA) at a significance level of p = 0.05. Where applicable, multiple comparisons were performed using Tukey’s post-hoc test.
| 3. Results and Discussion | ▴Top |
3.1. Quantification of Nitrogenous components
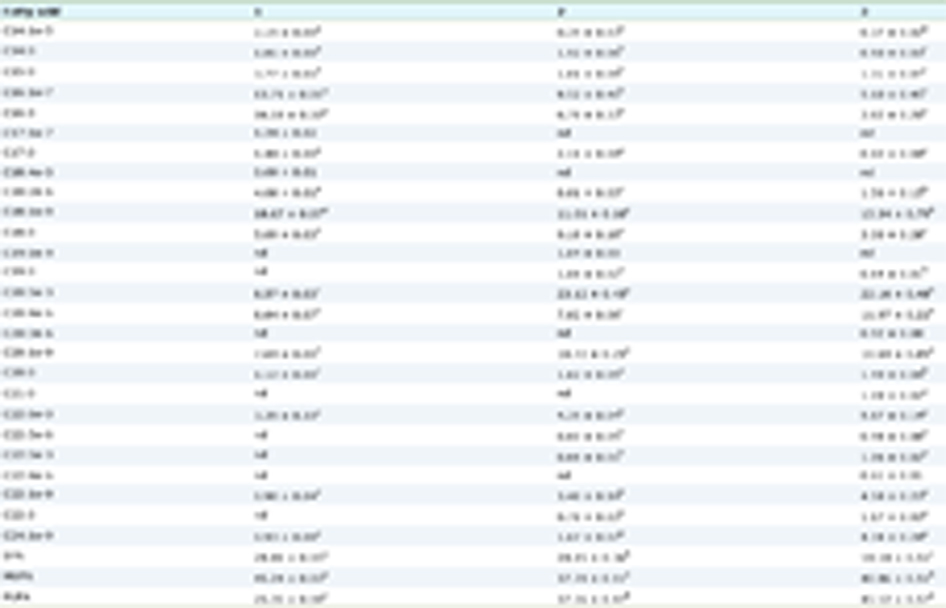
Kjeldahl analysis of P. purpureum and P. tricornutum revealed TN contents of 2.03 ± 0.02 and 2.86 ± 0.01% (w/w) dry weight, respectively (Table 1). Traditionally, a Nitrogen to protein (NTP) conversion factor of 6.25 is used to estimate the protein contents of different plant and animal samples from experimentally obtained TN values.For the purposes of this study, the NTP conversion factor of 4.58 was used as it was deemed suitable for all microalgal species, treatment and growth phases (Lourenco et al., 2004). An issue in the use of NTP conversion factors for quantification of protein content when estimated from TN, is that this approach may overestimate the actual protein content as it assumes that all nitrogen in the sample is PN and does not take into account the nitrogen present in DNA, RNA, mycosporine-like amino acids and intercellular nitrogen stores, i.e., the non-protein nitrogen (NPN) (Lourenco et al., 1998; Fujihara et al., 2001; Lourenco et al., 2004). Here, the organic reagent TCA was used to precipitate the proteinaceous material in the microalgal samples, which was then separated from the NPN containing fraction by centrifugation. Kjeldahl analysis of these two fractions revealed NPN values of 0.47 ± 0.08 and 1.02 ± 0.02% (w/w) and corresponding PN values of 1.50 ± 0.04 and 1.93 ± 0.02% (w/w) for P. purpureum and P. Tricornutum, respectively (Table 1). Applying the NTP conversion factor of 4.58 (Lourenco et al., 2004)to the PN values obtained revealed protein contents of 6.87 and 8.85% (w/w)for P. purpureum and P. tricornutum, respectively. These values arelower than the previously reported protein contents for each of these species ranging from 44.56 to 54.66% (Fernández-Reiriz et al., 1989; Becker, 1994), which could be demonstrative of how protein contents of microalgae are overestimated when an NTP factor of 6.25 is used with the experimentally determined TN values of biomass.
 Click to view | Table 1. Total nitrogen (TN), non-protein nitrogen (NPN) and protein nitrogen (PN) contents of Porphyridium purpureum and Phaeodactylum tricornutum |
3.2. SDS-PAGE profiles of P. purpureum and P. tricornutum
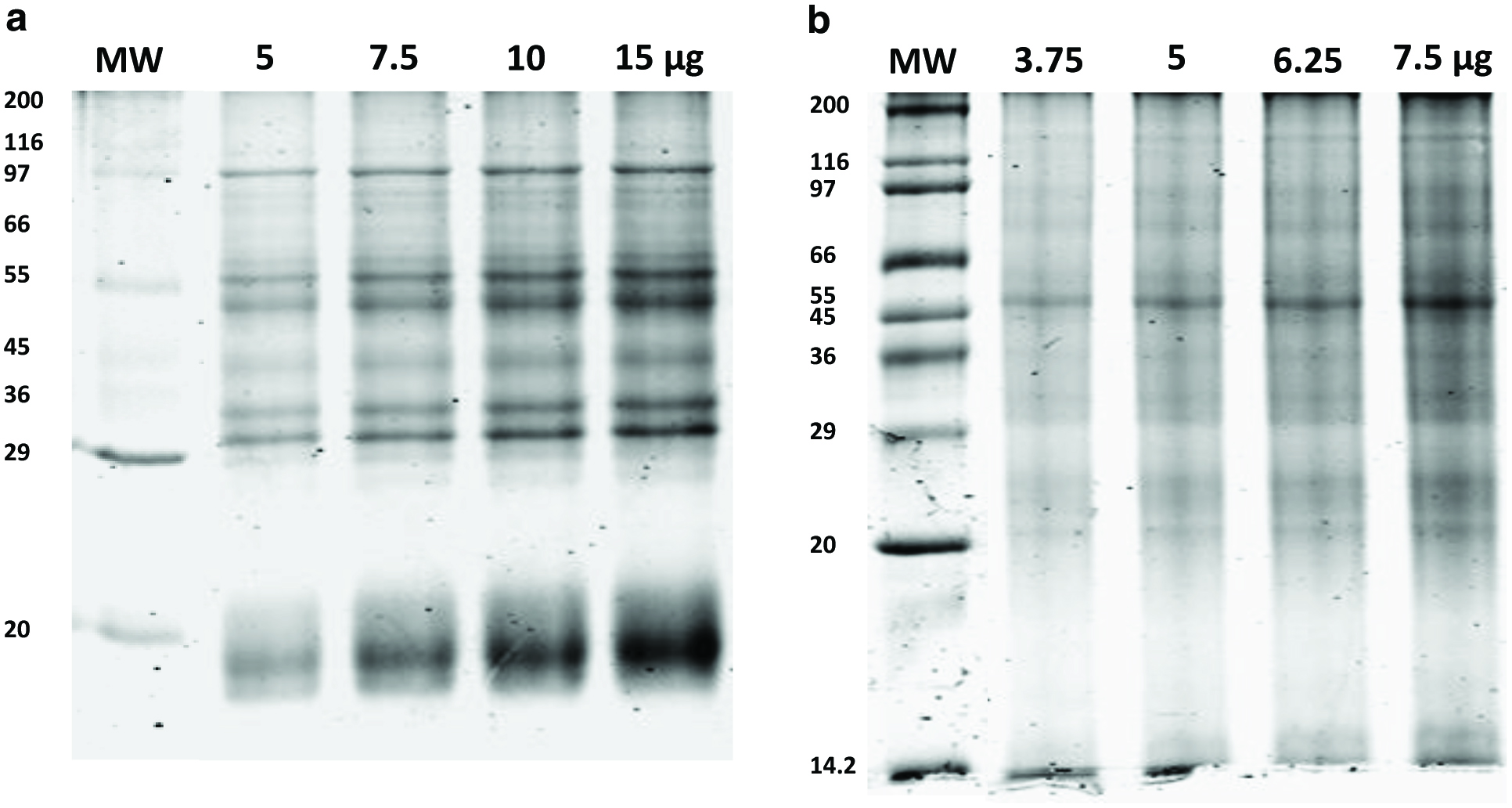
Figure 1 shows the SDS-PAGE profiles of the total proteins extracted from milled freeze-dried P. purpureum and P. tricornutum. The profile for P. purpureum comprised approximately 16 distinct bands ranging in mass from 15 to 200 kDa, some of which are only visible when 15 μg protein was loaded on to the gel (Fig. 1a). Particularly prominent bands visible across all lanes are those resolving at 30, 32, 45, 55, 57 and 95 kDa. There was also a very intense area of staining at 15-20 kDa. The identity of the majority of these proteins is unknown; however, some of these bands may be components of the P. purpureum phycobilisomes (PBs). PBs mostly contain phycoerythrin, phycocyanin and allophycocyanin, and their component polypeptideshave been shown to have molecular weights of 13.3, 14, 15, 18.7, 19.5, 21, 24, 25.5, 30.5, 31.5, 32.5, 38, 49, 60 and 95 kDa (Redlinger and Gantt, 1981). No bands below 15 kDa were visible in this study, but this may reflect differences in SDS-PAGE methodology and the estimation of molecular masses of proteins (Redlinger and Gantt, 1981). The proteins in the15-20 kDa region were the most intense area of staining, and its smeared appearance probably reflects the presence of a number of different proteins, possibly including members of the P. purpureum light-harvesting complex (Bhattacharya et al., 2013).
 Click for large image | Figure 1. Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) profiles of proteins extracted from milled freeze-dried microalgae. |
The SDS-PAGE profile of P. Tricornutum comprised 10 clear bands visible at 15, 18, 27, 29, 32, 45, 55, 60 and 100 kDa along with other indistinct bands visible throughout the lanes (Fig. 1b), consistent with previous reports (Chen et al., 2014). Additional bands were not visible when greater amounts of protein were loaded on the gel (compare lanes 1 and 4, Fig. 1b).This profile was visibly different to that of P. purpureum, and the bands were less sharp than those observed for P. purpureum (Fig. 1a). This may indicate a higher level of lipids or carbohydrates present in these microalgae. Diatom light harvest complexes, referred to as fucoxanthin chlorophyll proteins, have molecular mass values ranging from 17 to 23 kDa (Devaki and Grossman, 1993; Buchel, 2003) and have been previously observed to run as a doublet (Guglielmi et al., 2005) as in Fig. 1b. The identity of the other proteins is unknown.
3.3. Optimisation of protein extraction
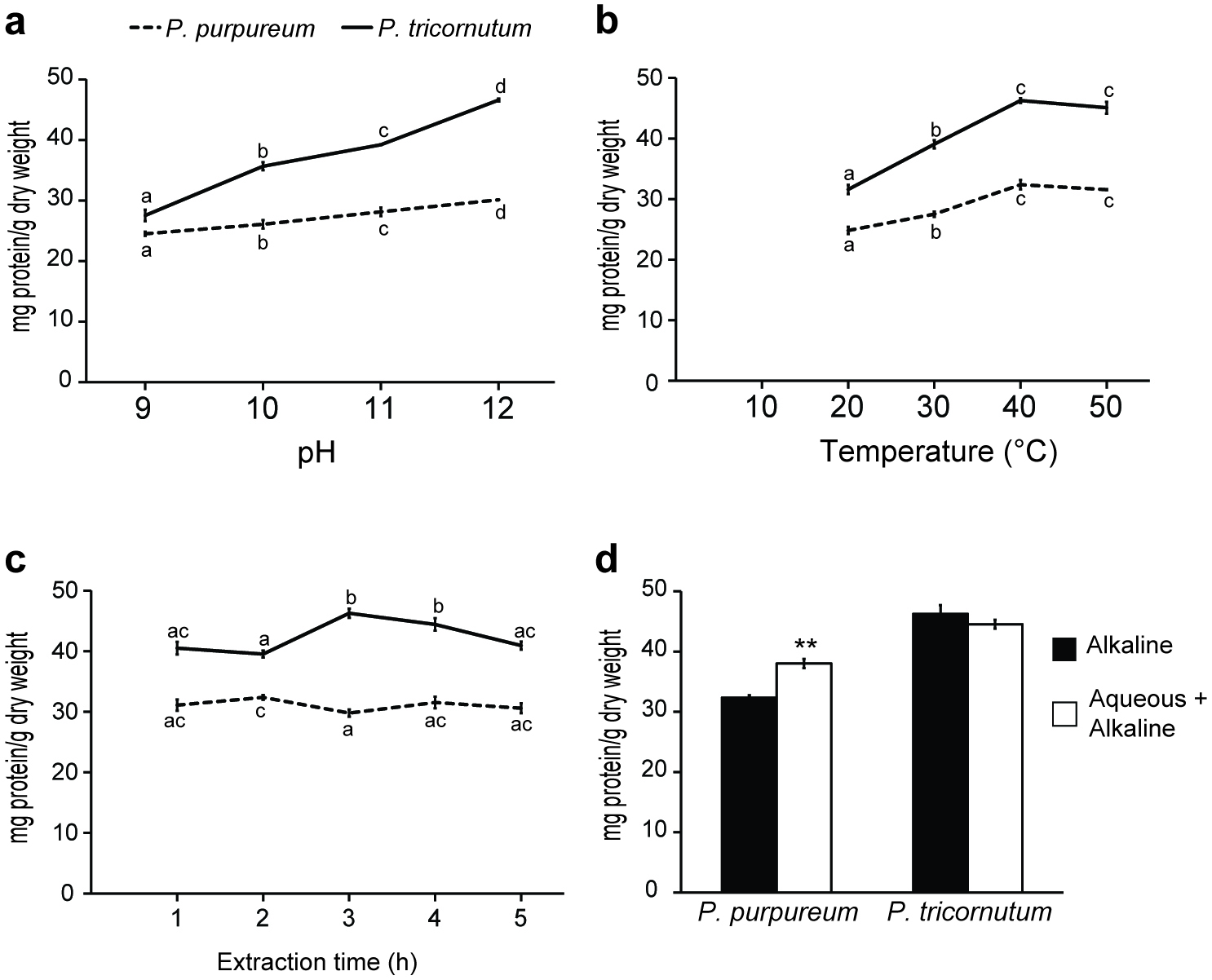
Figure 2 illustrates the effect of variation of pH, temperature and duration of extraction on the total protein recovery from milled freeze-dried microalgae. Alkaline solutions (NaOH) have been demonstrated to effectively solubilise and aid in the extraction of highly water insoluble proteins from macroalgae and microalgae (Fleurence et al., 1995; Barbarino and Lourenço, 2005). Previous studies have shown that increasing the pH of the alkaline solutions used for protein extraction has a beneficial effect on the total amount of protein recovered (Harnedy and FitzGerald, 2013a). Figure 2a shows that a significantly higher amount of protein was extracted at pH 12 than at pH 9, 10 or 11 for both P. purpureum and P. tricornutum. The amount of protein recovered from P. purpureum increased from 24.52 ± 0.33 to 30.12 ± 0.23 mg/g of dried biomass, and a similar effect was observed for P. tricornutum with yield increasing from 27.54 ± 0.95 to 46.59 ± 0.24 mg/g of dried biomass (Fig. 2a) when pH was increased from 9 to 12. Therefore, pH 12 was chosen for all further extraction studies.
 Click for large image | Figure 2. Effects of various factors on the recovery of alkaline soluble protein from milled freeze-dried Porphyridium purpureum and Phaeodactylum tricornutum. |
Figure 2b demonstrates that as the temperature was increased from 20 to 40 °C, the amount of total protein recovered from both species increased significantly. The amount of protein recovered from P. purpureum increased from 24.82 ± 0.75 to 32.37 ± 0.39 mg/g of dried biomass, and a similar effect was observed for P. tricornutum with yield increasing from 31.59 ± 0.57 to 46.27 ± 0.77 mg/g of dried biomass. Further increasing the temperature from 40 to 50 °C did not significantly increase the protein recovery from either species (Fig. 2b). An extraction temperature of 40 °C was therefore selected for further studies. This temperature has previously been used as the optimal to extract proteins from microalgae in alkaline solution (Safi et al., 2013; Safi et al., 2014). Interestingly, studies examining protein extraction from the red macroalga, Palmaria palmata, reported no significant increase in the mean concentration of protein recovered as the temperature was increased from room temperature to 50 °C (Harnedy and FitzGerald, 2013a).
Previous protein extraction studies with algae reported conflicting results on the benefit of extended extraction times. Harnedy and Fitzgerald (2013a) found no increase in the concentration of protein recovered from P. palmata when the duration of extraction was increased from 0.5 to 3.0 h. However, a major increase in the yield of protein extracted from the microalga Scenedesmus acutus was observed with increasing extraction time (Venkataraman and Shivashankar, 1979). Here, Fig. 2c shows that increasing the duration of alkaline extraction for P. purpureum from 1 to 5 h did not significantly increase or decrease the amount of protein recovered at the 1 h time point (31.09 ± 0.91 mg/g of dried biomass). In the case of P. tricornutum, Fig. 2c demonstrates that as the extraction time was increased from 1 to 3 h, the quantity of total protein recovered increased significantly from 40.49 ± 1.05 to 46.27 ± 0.77 mg/g of dried biomass. Further increases in extraction time did not positively affect protein yield, and actually led to a decrease in protein recovery. Alkaline extraction times of 2 h and 3 h were chosen for further studies withP. purpureum and P. tricornutum, respectively.
Fleurence et al. (1995) showed that performing an aqueous extraction prior to alkaline extraction increased the yield of total protein extracted from the edible seaweeds Ulva rigida and Ulva rotundata. This was also the case for P. purpureum, but not P. tricornutum herein. Figure 2d shows that a prior aqueous extraction (overnight at 4 °C) from P. purpureum significantly increased the recovery of protein from 32.37 ± 0.39 to 38.03 ± 1.44 mg/g of dried biomass. However, Fig. 2d also shows that performing an alkaline extraction alone leads to the recovery of a higher amount of protein (46.27 ± 0.77 mg/g of dried biomass) from P. tricornutum than combining alkaline and aqueous treatments (44.54 ± 0.72 mg/g of dried biomass). These data indicate that an overnight aqueous extraction prior to alkaline extraction was beneficial for optimal protein recovery from P. purpureum but not for P. tricornutum.
The extraction conditions used for P. purpureum were overnight aqueous extraction at 4 °C, followed by 2 h incubation with agitation at pH 12, 40 °C. Similar conditions were found to be optimal for maximal protein recovery from P. tricornutum, but without the prior aqueous extraction step and with the longer period of alkaline extraction of 3 h.
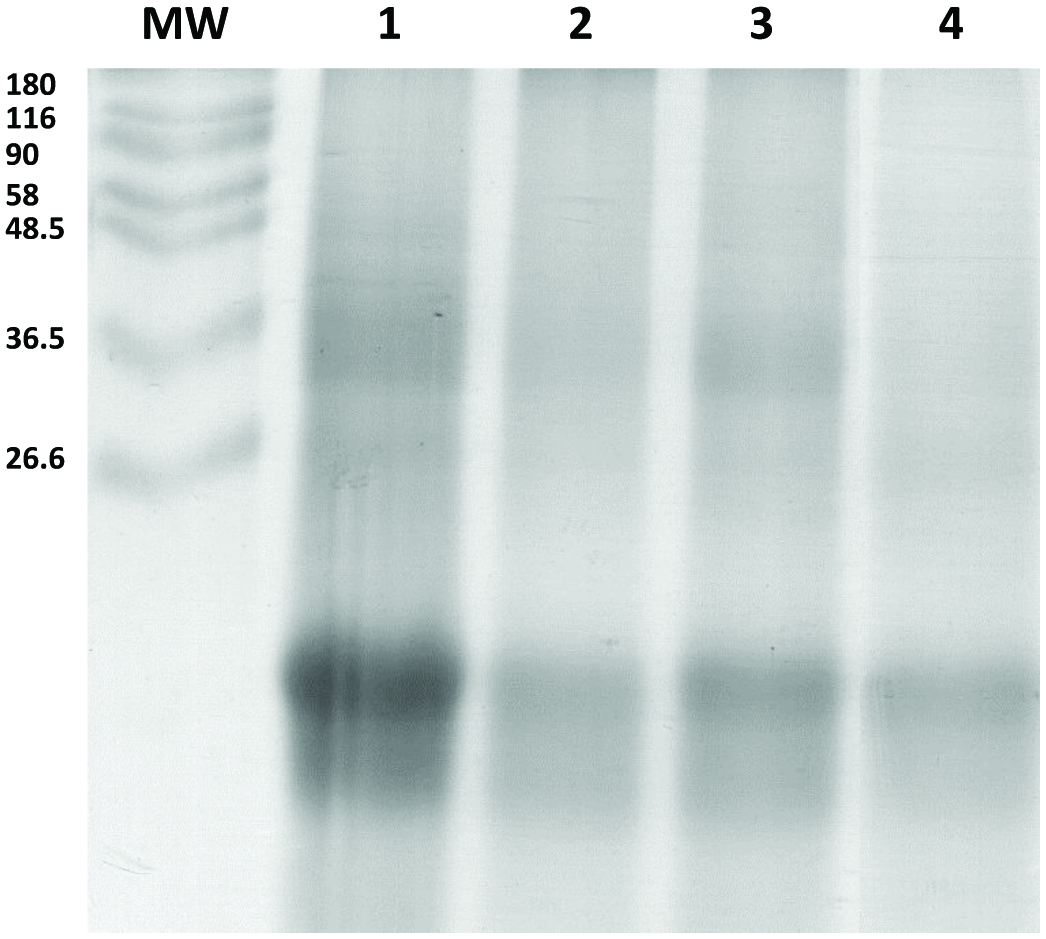
Figure 3, lanes 1 to 3, shows the SDS-PAGE profile of the aqueous- and alkaline-soluble proteins obtained from P. purpureum. Compared to the alkaline soluble protein extract (lane 2), where 2 main protein bands at 18 and 20 kDa and an indistinct smear of higher molecular weight proteins were visible, the aqueous soluble protein extract had a greater number of protein bands ranging in size from 18 to 58 kDa (lane 1). The most prominent area of staining was observed between 18 to 20 kDa in both cases, and when the fractions are combined (lane 3). Figure 3, lane 4, shows the SDS-PAGE profile of alkaline soluble proteins obtained from P. tricornutum. A clear band was visible at 18 kDa, but the higher molecular weight proteins are not visible as individual bands, but rather as a smear of proteins. In comparison to the SDS-PAGE images in Fig. 1, the band patterns in Fig. 3 are much more indistinct with a lot of smearing. This may reflect the presence of contaminants such as carbohydrates and lipids in the aqueous- and alkaline-soluble extracts which are removed during the sequential precipitations involved in the preparation of samples for SDS-PAGE profiling of whole microalgae.
 Click for large image | Figure 3. SDS-PAGE profiles of Porphyridium purpureum and Phaeodactylum tricornutum protein extracts. |
3.4. Endoproteinase activity
The aim of this study was to extract intact proteins from microalgae to generate hydrolysates for investigation of bioactivity. Therefore, it was important to establish if the extracts contain endogenous microalgal protein degrading enzymes. The presence of such endoproteinases would have an effect on the methods and conditions utilised for protein extraction from the biomass. The endoproteinase activities of protein fractions from both microalgae strains were tested at 22 and 40 °C to reflect experimental extraction conditions and also to examine stability at room temperature. The P. purpureum aqueous extract does not possess endogenous proteolytic activity following incubation at either 22 or 40 °C (Table 2). Therefore, the overnight aqueous extraction may be performed at room temperature without protein degradation. The P. purpureum alkaline-soluble extract possessed low to negligible endoproteinase activity on incubation at 22 and 40 °C, respectively. This indicates that self-hydrolysis of alkaline soluble proteins will not occur under the extraction conditions chosen. The alkali extract from P. tricornutum possesses some endoproteinase activity at 40 °C (Table 2), which may explain why a small reduction in the amount of total protein recovered from P. tricornutum was observed during alkaline extraction of more than 3 h in duration (Fig. 3c).
 Click to view | Table 2. Endoproteinase activity in Porphyridium purpureum and Phaeodactylum tricornutum protein extracts |
3.5. Characterisation of hydrolysates
Hydrolysis of protein extracts from edible macroalgae using the food-grade proteolytic preparations, Alcalase 2.4 L and Flavorzyme 500 L, leads to the generation of bioactive peptides as previously described by Harnedy and FitzGerald (2013b). Alcalase, a proteolytic preparation derived from Bacillus licheniformis mainly contains subtilisin endoproteinase and a minor glutamyl endopeptidase activity (Kalyankar et al., 2013).Flavourzyme, derived from Aspergillus oryzae, contains both endoproteinase and exopeptidase activity (Smyth and FitzGerald, 1998).Combined aqueous and alkaline protein extracts from P. purpureum and alkaline protein extracts from P. tricornutum were hydrolysed using an enzyme/substrate (E/S) ratio of 1% (v/w) of Alcalase and Flavourzyme for 4 h at 50 °C. A no-enzyme control sample was also incubated for 4 h at 50 °C. Figure 4a shows the changes in amino group concentration over 4 h in control (no-enzyme) and enzyme-treated P. purpureum and P. tricornutum protein extracts. An increase of 47.59 ± 1.68 and 21.12 ± 0.16 mg amino group/g of protein was observed during the hydrolysis of the protein extracts from P. purpureum and P. tricornutum, respectively. Interestingly, although protein extracts from both microalgal species were incubated with the same proteolytic preparations, the concentration of amino groups released was higher for P. purpureum than P. tricornutum. Additionally, no significant changes were observed in the amino group concentration during 4 h incubationin the control samples for both microalgal species (Fig. 4). Therefore, the increase of amino group concentration during the hydrolysis process is attributable to the proteolytic action of Alcalase and Flavourzyme alone. The small reduction in the amount of total protein recovered from P. tricornutum observed after more than 3 h alkaline extraction (Fig. 2c) may be due to enzymes present in the protein extracts which may have been removed during the isoelectric precipitation protein-enrichment process.
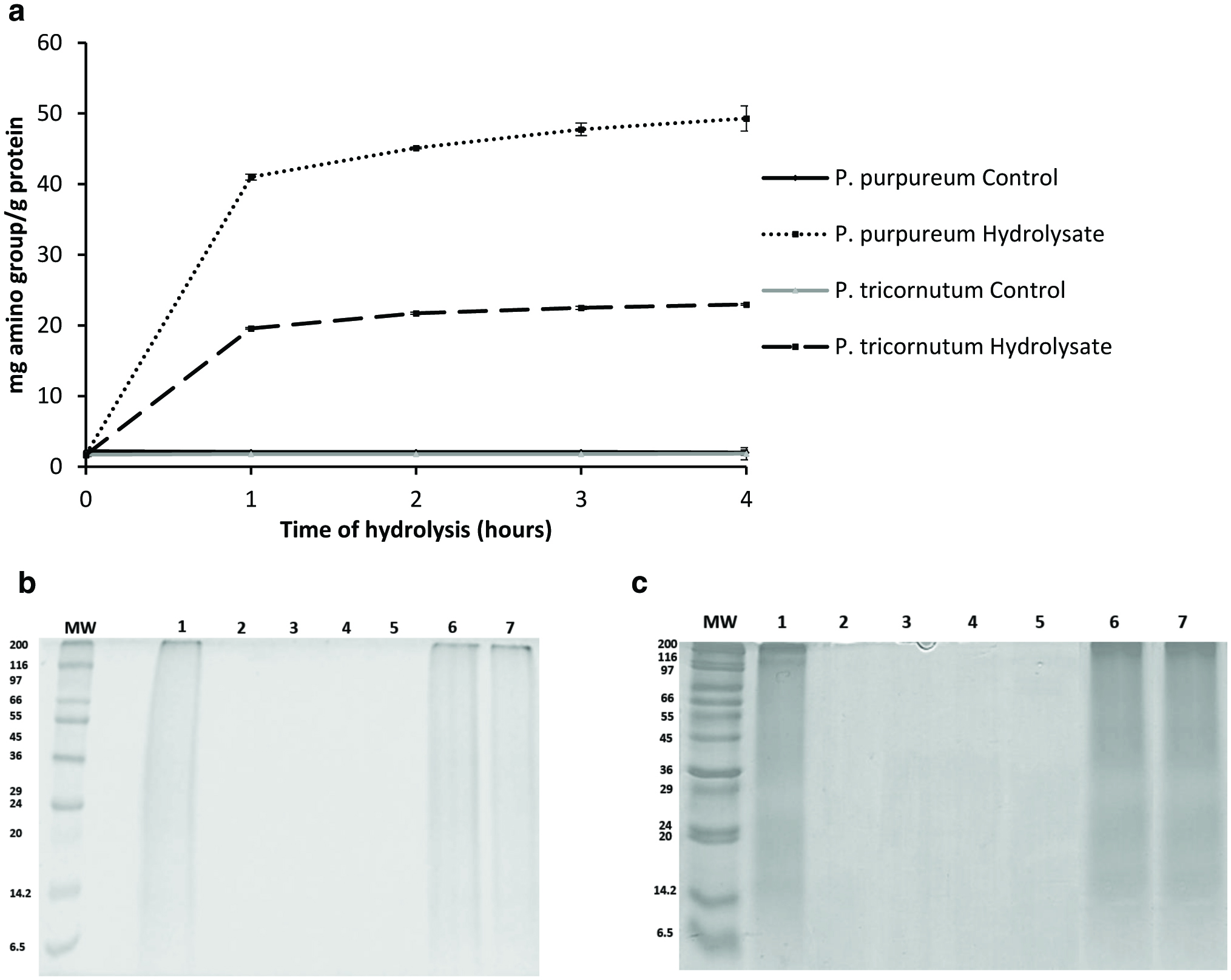 Click for large image | Figure 4. Monitoring the hydrolysis of Porphyridium purpureum and Phaeodactylum tricornutum protein extracts. |
The hydrolysis effects of Alcalase and Flavourzyme were further confirmed by SDS-PAGE (Fig. 4b, c) and GP-HPLC analysis (Fig. 5). SDS-PAGE was used to visualise the total protein profiles of the unhydrolysed substrates and their degradation during incubation with the proteolytic enzyme preparations. The SDS-PAGE profiles (Fig. 4b and c) indicatethat the protein extracts from both microalgae were significantly hydrolysed by Alcalase and Flavourzyme even after 1 h incubation(Fig. 4b [lane 2], c [lane 2]). However, the same protein extracts were not degraded when incubated without enzymes at 50 °C for 4 h (Fig. 4b [lane 7] and c [lane 7]). These SDS-PAGE profiles do not contain distinct protein bands, but nonetheless clearly demonstrate that the protein extracts are hydrolysed by the proteolytic preparations, Alcalase and Flavourzyme.
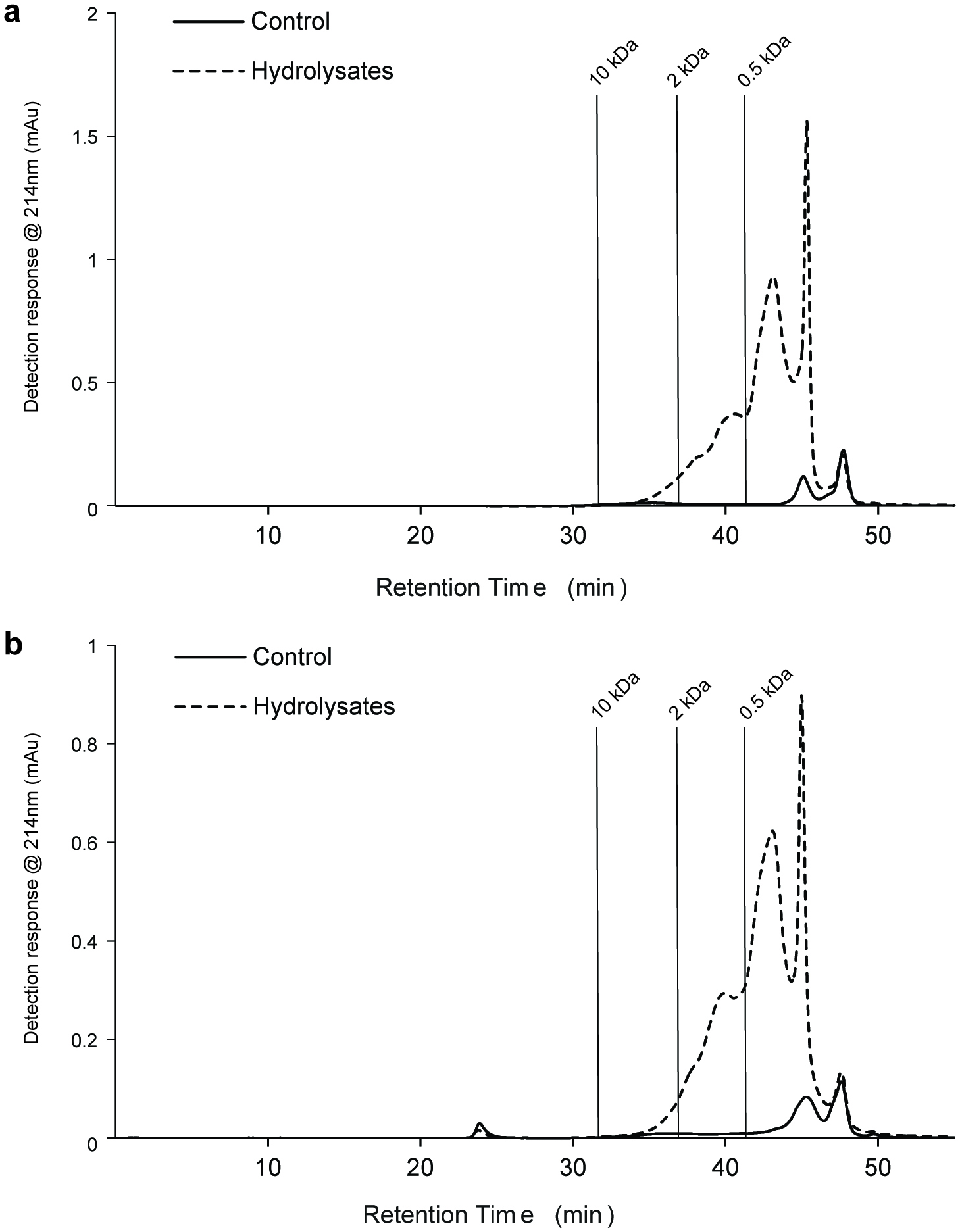 Click for large image | Figure 5. Gel permeation high performance liquid chromatography (GP-HPLC) profiles showing the molecular mass distribution of the soluble proteinaceous components in Porphyridium purpureum and Phaeodactylum tricornutumprotein hydrolysates after 4 h hydrolysis. |
GP-HPLC analysis was used to assess the molecular weight distribution of soluble proteinaceous components in the unhydrolysed protein fractions and associated hydrolysates (Fig. 5). The proteolytic enzymeshad a significant effect on the molecular masses of the resultant peptides generated, with the quantity of low molecular weight peptides (<10 kDa) being higher in the protein hydrolysates generated with Alcalase and Flavourzyme than in the no enzyme control for both P. purpureum (Fig. 5a) and P. tricornutum (Fig. 5b). Most of these peptides were <0.5 kDa. Low molecular mass peptides are generally more readily absorbed across the gastrointestinal tract(Meisel et al., 2005). Therefore, if bioactive, these <0.5 kDa microalgae-derived peptides could potentially reach their target site in a bioavailable format.
3.6. DPP-IV Inhibition
DPP-IV inhibitors reduce DPP-IV activity and increase the lifetime of the incretins, GLP-1 and GIP, and maybe used to control T2DM. Many natural dietary protein-derived DPP-IV-inhibitory peptides from macroalgae (Harnedy and FitzGerald, 2013b), fish (Huang et al., 2012), and milk (Lacroix and Li-Chan, 2013; Nongonierma et al., 2017)proteins, have been reported.To our knowledge, no information regarding the ability of microalgal-derived protein hydrolysates to inhibit DPP IV activity has been reported to date.
The protein hydrolysates from P. purpureumand P. tricornutumhad significantly more potent DPP-IV inhibitory activity compared to the unhydrolysed control samples (Table 3). P. purpureum protein extracts hydrolysed with Alcalase and Flavourzyme had an IC50 value of 2.28 ± 0.21 mg/mL, whereas the control protein sample had an IC50 value of 5.07 ± 0.21 mg/mL. P. tricornutum protein hydrolysates had an IC50 value of 2.67 ± 0.19 mg/mL, which is significantly different from that observed for the unhydrolysed control samples (IC50 value of 3.34 ± 0.26 mg/mL). To our knowledge, this is the first report of microalgal protein hydrolysates having DPP-IV inhibitory activity and theseIC50 values are comparable to previously published food-derived DPP-IV inhibitory protein hydrolysates. Hatanaka et al. (2012) reportedrice bran peptides produced with Umamizyme G possessingan IC50 value of 2.3 ± 0.1 mg/mL, and Atlantic salmon skin-derived collagen hydrolysates achieved 15–45.2% inhibition of DPP-IV when assessed at 5 mg/mL(Li-Chan et al., 2012). Alaska pollock skin-derived collagen hydrolysates had IC50 values in the range 0.80-2.59 mg/mL (Guo et al., 2015), and P. palmata hydrolysates had IC50 values in the range 1.65–4.60 mg/mL (Harnedy and FitzGerald, 2013b). Interestingly, the hydrolysates derived from Alaska pollock skin-derived collagen and P. palmata displayed different degrees of DPP-IV inhibitory activity depending on the proteolytic preparation used (Harnedy and FitzGerald, 2013b; Guo et al., 2015), suggesting that the potency of the P. purpureumand P. tricornutum DPP-IV inhibition could perhaps be improved by using different enzymes.
 Click to view | Table 3. Dipeptidyl peptidase (DPP) IV inhibitory activity, ferric reducing antioxidant power (FRAP) and oxygen radical antioxidant capacity (ORAC) of Porphyridium purpureum and Phaeodactylum tricornutum protein hydrolysates |
3.7. Antioxidant activity
Oxidative stress plays a key role in the pathogenesis of T2DM complications including atherosclerosis, retinopathy, and nephropathy (Rochette et al., 2014; Oseguera-Toledo et al., 2015). Therefore, foods containing multifunctional bioactive peptides could be extremely useful for the prevention and management of T2DM and its associated complications. Due to their predominantly photoautotrophic life under variable environmental conditions, microalgae may encounter oxidative and free radical stress, which are considered to have led to evolution of efficient protective mechanisms against oxidative damage (Becker, 1994).P. purpureum and P. tricornutum Alcalase/Flavourzyme hydrolysates have significantly higher antioxidant activity (FRAP and ORAC) compared to the unhydrolysed control samples (Table 3).The ORAC value obtained for theP. purpureumhydrolysate (478.94 ± 34.43 μmol TE/g FDP) was significantly higher than that seen for its unhydrolysed no-enzyme control (143.58 ± 11.33 μmol TE/g FDP). By comparison the P. tricornutum hydrolysate yielded a lower ORAC value (155.74 ± 38.30 μmol TE/g FDP). Overall, these values are comparable to, if not higher, than those reported for P. palmate (Harnedy and FitzGerald, 2013b) and other marine protein hydrolysates (Khantaphant et al., 2011; Cian et al., 2012). The results from the in vitro ORAC assay cannot be extrapolated to potential antioxidant effects in vivo (USDA, 2010; Cömert and Gökmen, 2018). However, it is known that antioxidant compounds in foods can have other functions besides their ability to scavenge free radicals. Therefore, at least two different assays are generally used to assess the antioxidant activity of food derived compounds(Cumby et al., 2008; Alzahrani et al., 2018).
The metal chelating activity, i.e., the ferric reducing capacity (FRAP) of the hydrolysates was also determined herein. FRAP values of 13.97 ± 0.97 and 15.04 ± 0.54 μmol TE/g FDP were respectively recorded for the hydrolysates generated from the P. purpureum and P. tricornutum protein extracts (Table 3). These values were significantly higher than the FRAP values of 3.16 ± 0.35 and 1.69 ± 0.43 μmol TE/g FDP obtained for the P. purpureum and P. tricornutum unhydrolysed no-enzyme controls, respectively.
The antioxidant properties of peptides are highly influenced by molecular mass and molecular structure properties (Suetsuna et al., 2000). TNBS analysis of the microalgal Alcalase/Flavourzyme hydrolysates showed that the concentration of amino groups released was higher for P. purpureum than P. tricornutum (Fig. 4a). It is possible that using a different proteolytic enzyme or combination of enzymes could lead to P. tricornutum hydrolysates with higher antioxidant activities as demonstrated by other studies (Harnedy and FitzGerald, 2013b; Beaulieu et al., 2016). Bondu et al. (2015) indicated that most of antioxidant peptides from the macroalgae Solieria chordalis hydrolysates came from ribulose-1, 5-biphosphate carboxylase/oxygenase (RiBiSCo). It is also possible that further beneficial effects may be observed if different microalgal protein hydrolysates were combined. GP-HPLC analysis showed that the vast majority of microalgal peptides generated with Alcalase/Flavourzyme treatment were <0.5 kDa (Fig. 5). This is in agreement with published studies which report that peptides exhibiting antioxidant activity generally have low molecular masses (Pihlanto, 2006). Interestingly, low molecular weight peptides are more likely to be better absorbed in the intestine (Meisel et al., 2005), making these peptides an interesting prospect for investigation as functional food ingredients. These hydrolysates may also find application for use as food preservatives to combat lipid peroxidation and prolong food quality.
Due to the limited quantity of sample available it was not possible to perform amino acid compositional analysis. However, previous reports on the amino acid composition of P. tricornutum and P. purpureum indicated that they had essential amino acid contents equivalent to 53.5 and 36.6% of total amino acids, respectively (Heaney-Kieras et al., 1977; Brown, 1991). However, it should be noted that Heaney-Kieras et al. (1977) did not report values for Arg, His, Met, Pro, Trp and Cys. Future work will involve determination of an amino acid composition profile for the proteins extracted from the microalgae studied herein.
| 4. Conclusion | ▴Top |
This study reports an alkaline protein extraction method for P. purpureum and P. tricornutum and demonstratesfor the first time that Alcalase/Flavourzyme protein hydrolysates generated from these species had DPP-IV inhibitory and antioxidantactivities comparable to, or more potent than, other published marine protein hydrolysates. These data clearly indicate that protein hydrolysates from P. purpureum and P. tricornutum havepotential as multifunctional antioxidant and DPP-IV inhibitory functional food ingredients. It will be necessary to perform sequence analysis to identify the specific DPP-IV inhibitory and anti-oxidant peptides, as well as in vivo studies to validate their physiological effectsfor further development of these hydrolysates as functional food ingredients.
DPP-IV, dipeptidyl peptidase-IV; EPA, eicosapentaenoic acid;FDP, Freeze-dried powder; FRAP, Ferric reducing antioxidant power; GIP, glucose-dependent insulinotropic polypeptide; GLP-1, glucagon-like peptide 1;GP-HPLC, Gel permeation high performance liquid chromatography;h, hour/s; min, minute/s;IC50(inhibitory concentration that inhibits enzyme activity by 50%; NPN, non-protein Nitrogen; NTP, nitrogen to protein;PN, protein Nitrogen; ORAC, Oxygen radical absorbance capacity;P. purpureum, Porphyridium purpureum; P. tricornutum(Phaeodactylum tricornutum; SDS-PAGE, sodium dodecyl sulphate – polyacrylamide gel electrophoresis; T2DM, Type 2 diabetes mellitus; TCA, trichloroacetic acid; TE, Trolox Equivalent; TN, total Nitrogen; TNBS, trinitrobenzenesulphonic acid; WHO, World Health Organisation
Acknowledgments
The authors would like to acknowledge Pádraigín Harnedy for advice and helpful discussions, and Martina O’Keefe for assistance with GP-HPLC analysis. Tara Flaherty is also acknowledged for contributions on the quantification of NPN and PN of algal samples.
This work was supported under the National Development Plan 2007-2013, through NutraMara, the Marine Functional Food Research Initiative, and the Food Institutional Research Measure, administered by the Department of Agriculture, Food, and the Marine, Ireland under grant issue 13/F/536.
| References | ▴Top |