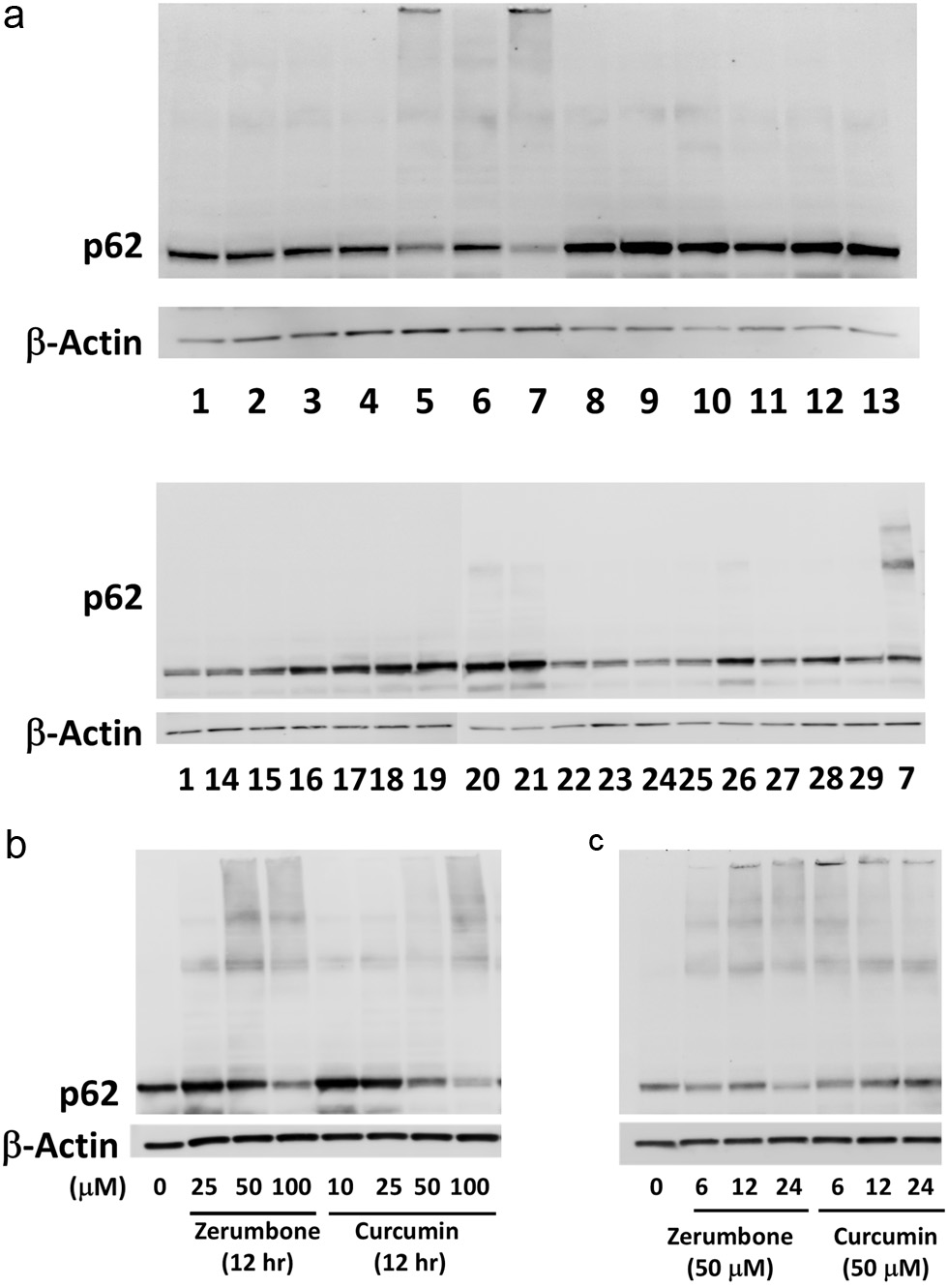
Figure 1. (a) Curcumin (lane 5) and zerumbone (lane 7) markedly formed the p62 conjugated proteins. Hepa1c1c7 cells were treated with vehicle (0.5% DMSO) or phytochemicals for 12 h and then lysed for western blot analysis. β-actin was used as the internal standard. Lane 1, control; Lane 2, quercetin (100 μM); Lane 3, ellagic acid (50 μM); Lane 4, (–)-epigallocatechin-3-O-gallate (10 μM); Lane 5, curcumin (50 μM); Lane 6, resveratrol; Lane 7, zerumbone (100 μM); Lane 8, α-humulene; Lane 9, ursolic acid (5 μM); Lane 10, phenethyl isothiocyanate (10 μM); Lane 11, sulforaphane (25 μM); Lane 12, diallyl trisulfide c; Lane 13, benzoic acid (100 μM); Lane 14, fructose (5 mM): Lane 15, glucose (5 mM); Lane 16, L-arginine (5 mM); Lane 17, L-asparagine (1 mM); Lane 18, vitamin A (50 μM); Lane 19, vitamin D (50 μM); Lane 20, vitamin E (250 μM); Lane 21, vitamin K (500 μM); Lane 22, vitamin B1 (10 mM); Lane 23, vitamin B3 (niacin, 5 mM); Lane 24, vitamin B6 (10 mM); Lane 25, vitamin B12 (100 μM); Lane 26, vitamin C (100 μM); Lane 27, vitamin M (folic acid, 500 μM); Lane 28, MnSO4 (25 μM); Lane 29, ZnCl2 (50 μM). Effects of concentrations and incubation times of zerumbone, curcumin and resveratrol on p62 conjugation. (b) Hepa1c1c7 cells were treated with zerumbone (25, 50, 100 μM), curcumin (10, 25, 50, 100 μM) or vehicle for 12 h. (c) Cells were treated with zerumbone, curcumin, resveratrol (50 μM) or vehicle for 6, 12, 18 h. Expression levels of p62 protein were analyzed by western blotting, and β-actin was used as the internal standard. CUR, curcumin; ZER, zerumbone.
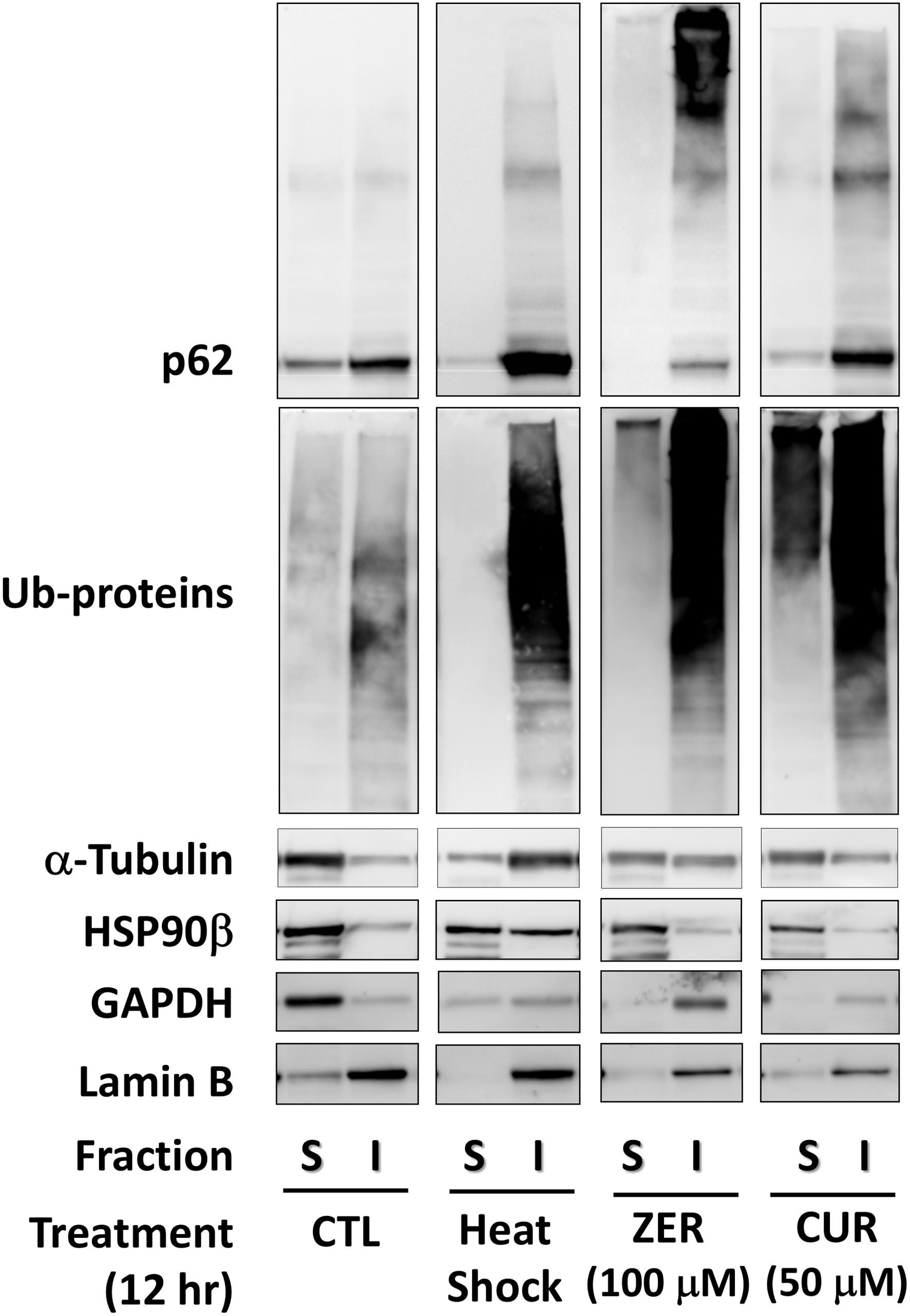
Figure 2. Insolubilization of housekeeping proteins by zerumbone and curcumin. Cells treated with zerumbone (100 μM), curcumin (50 μM) for 12 h were lysed, then fractionated into soluble and insoluble proteins using a ReadyPrep™ Protein Extraction Kit for western blot analysis. CTL, control; CUR, curcumin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HS, heat shock; I, insoluble; S, soluble; ZER, zerumbone.
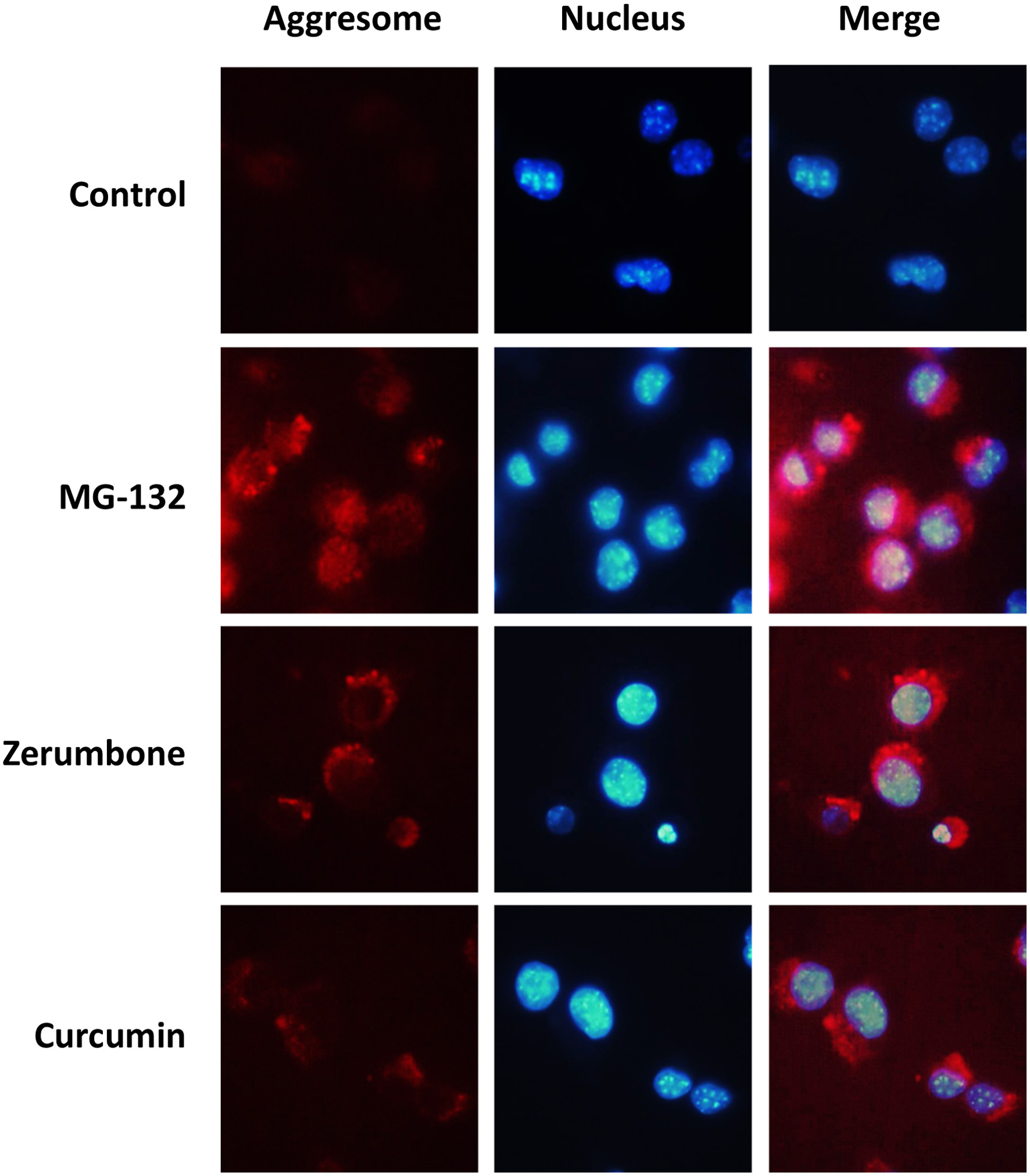
Figure 3. Detection of cellular aggresomes induced by phytochemicals. Hepa1c1c7 cells were treated with zerumbone (10 μM), curcumin (5 μM), or MG-132 (10 μM) for 12 h, and then cellular aggresomes and nucleus were stained using a ProteoStat Aggresome Detection Reagent and Hoechst 33342, respectively. CTL, control; CUR, curcumin; ZER, zerumbone.
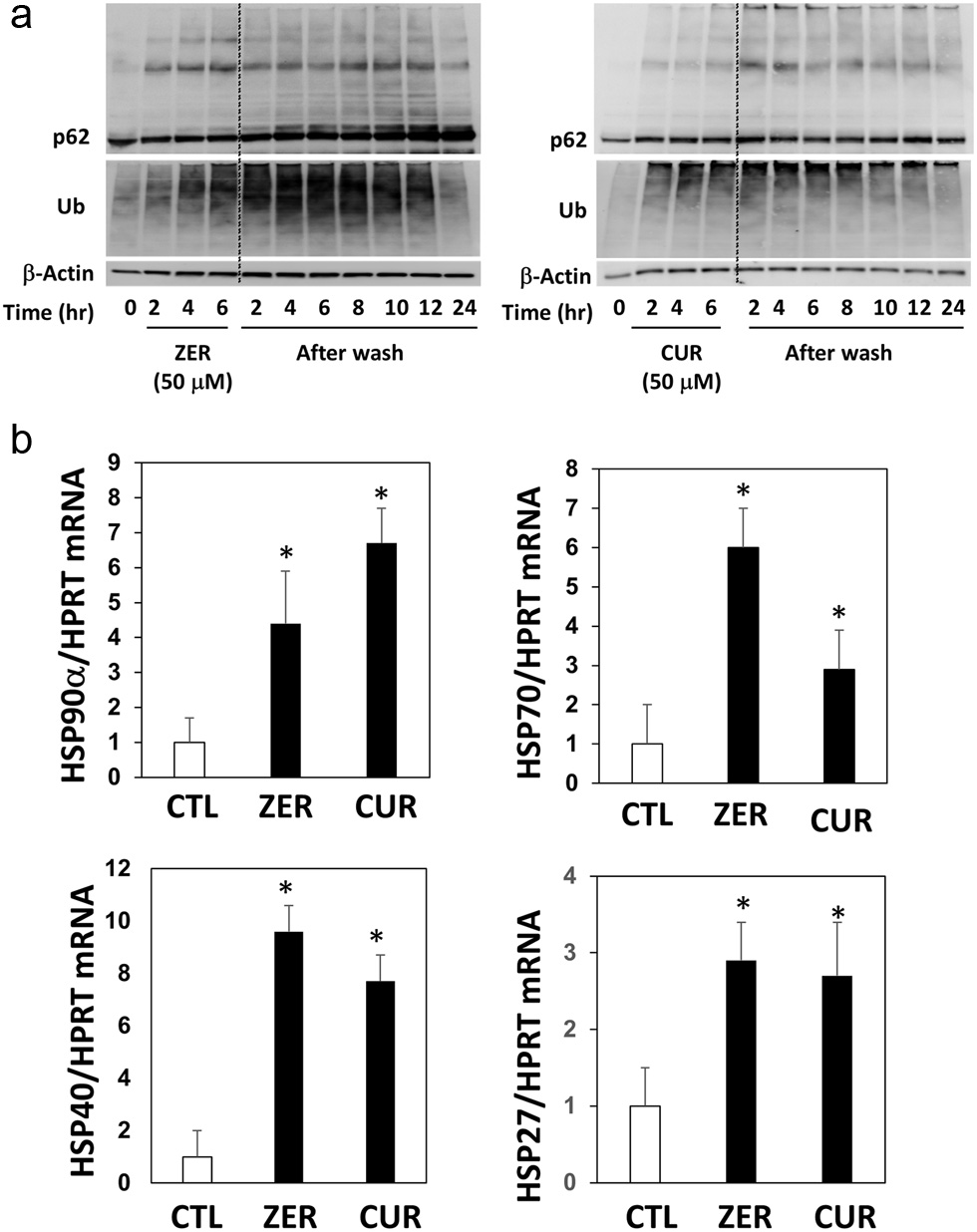
Figure 4. Time course of p62 conjugation and protein ubiquitination by zerumbone and curcumin after washing cells. Hepa1c1c7 cells were treated with (a) zerumbone, (b) curcumin (50 μM each), or vehicle control for 6 hours, then cultured in DMEM without FBS for another 2–24 h. The expression levels of p62 and ubiquitin were analyzed by western blotting. CUR, curcumin, zerumbone. (b) Hepa1c1c7 cells were seeded onto 24-well plates and treated with the sample or vehicle (0.5%, v/v) for 12 h. cDNA was synthesized using total RNA (1 µg) and real-time PCR was performed as follows: 50 °C 2 min and 95 °C 2 min for 1 cycle, and 95 °C for 15 sec, 60 °C for 30 sec, 72 °C for 30 sec for 99 cycles. The relative mRNA levels were estimated using the ΔΔCt method. HPRT was used as an internal standard. *P < 0.05 vs. CTL by Student’s t-test. CTL, control.
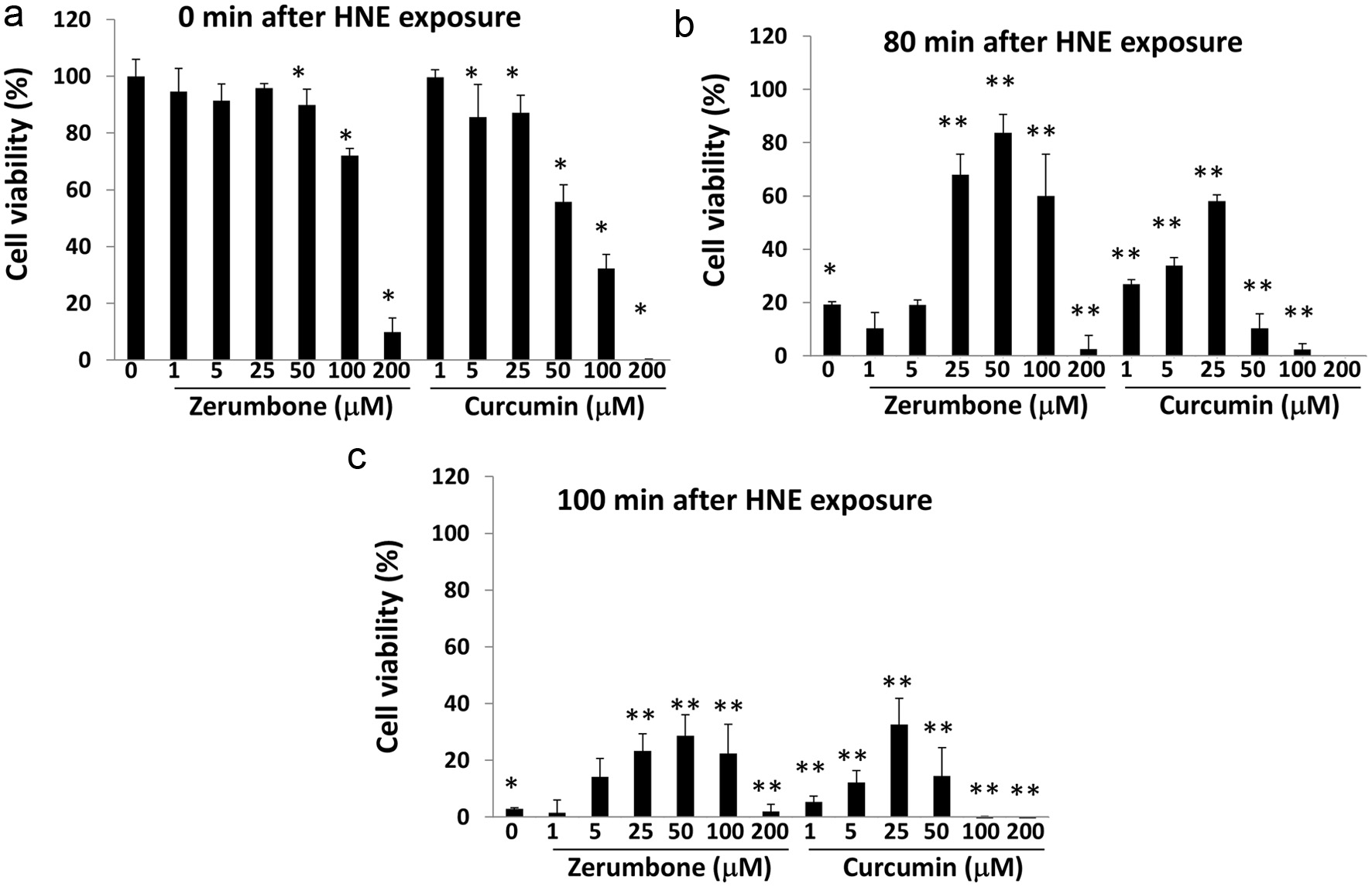
Figure 5. Zerumbone and curcumin conferred resistance to HNE-induced cell death. Hepa1c1c7 cells were pre-treated with each phytochemical (0–200 μM each) for 6 h. After washing, they were incubated in DMEM containing 10% FBS for another 18 hours, followed by exposure to HNE (0, 200 μM) for 0 (a), 80 (b), or 100 (c) min. Then, cell viability was determined using a WST-8 kit. Panel A, *P < 0.05 vs. CTL, Panel B, *P < 0.05 vs. CTL (0 min), **P < 0.05 vs. CTL (80 min). Panel C, *P < 0.05 vs. CTL (0 min), **P < 0.05 vs. CTL (100 min). CTL, control.




