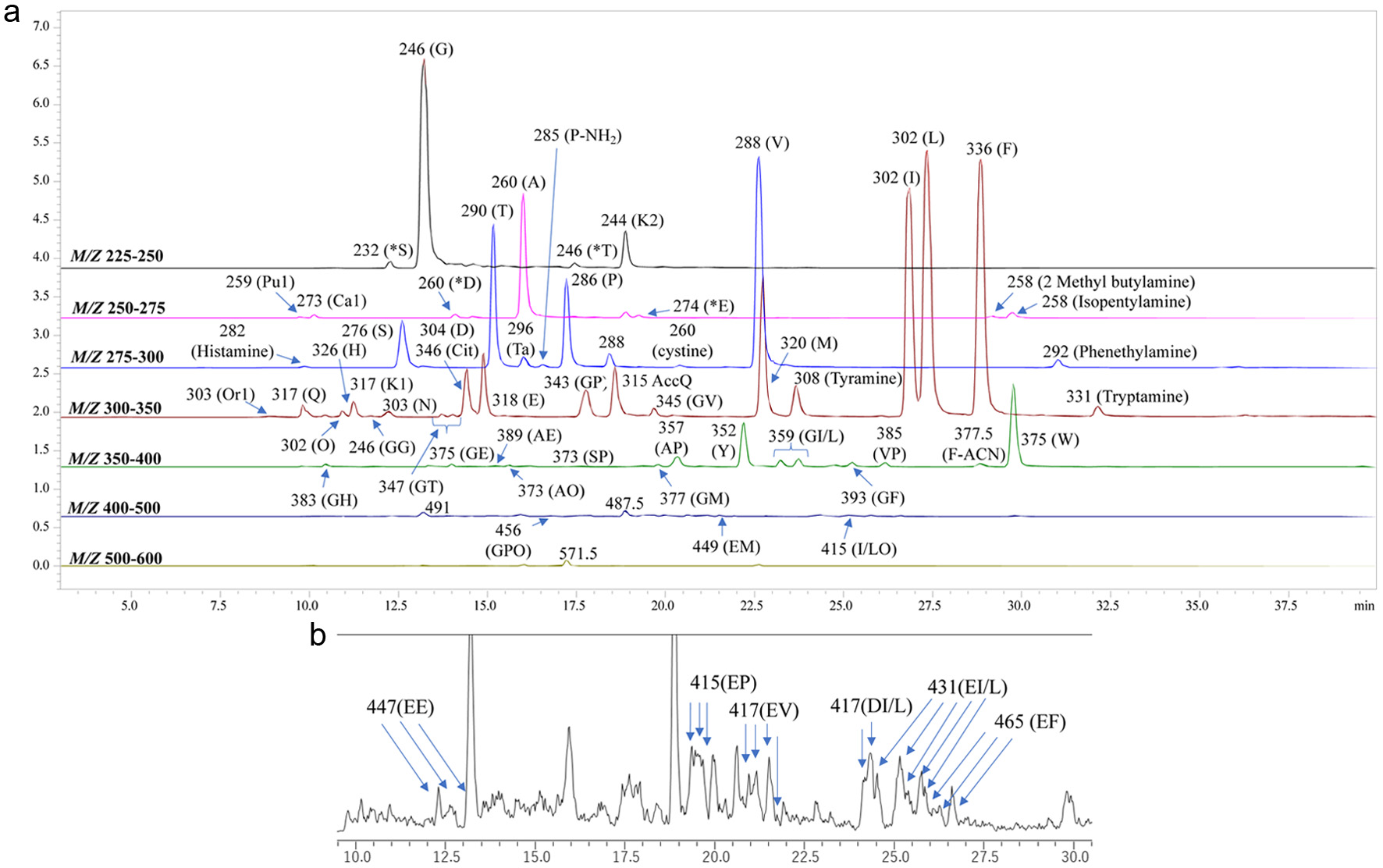
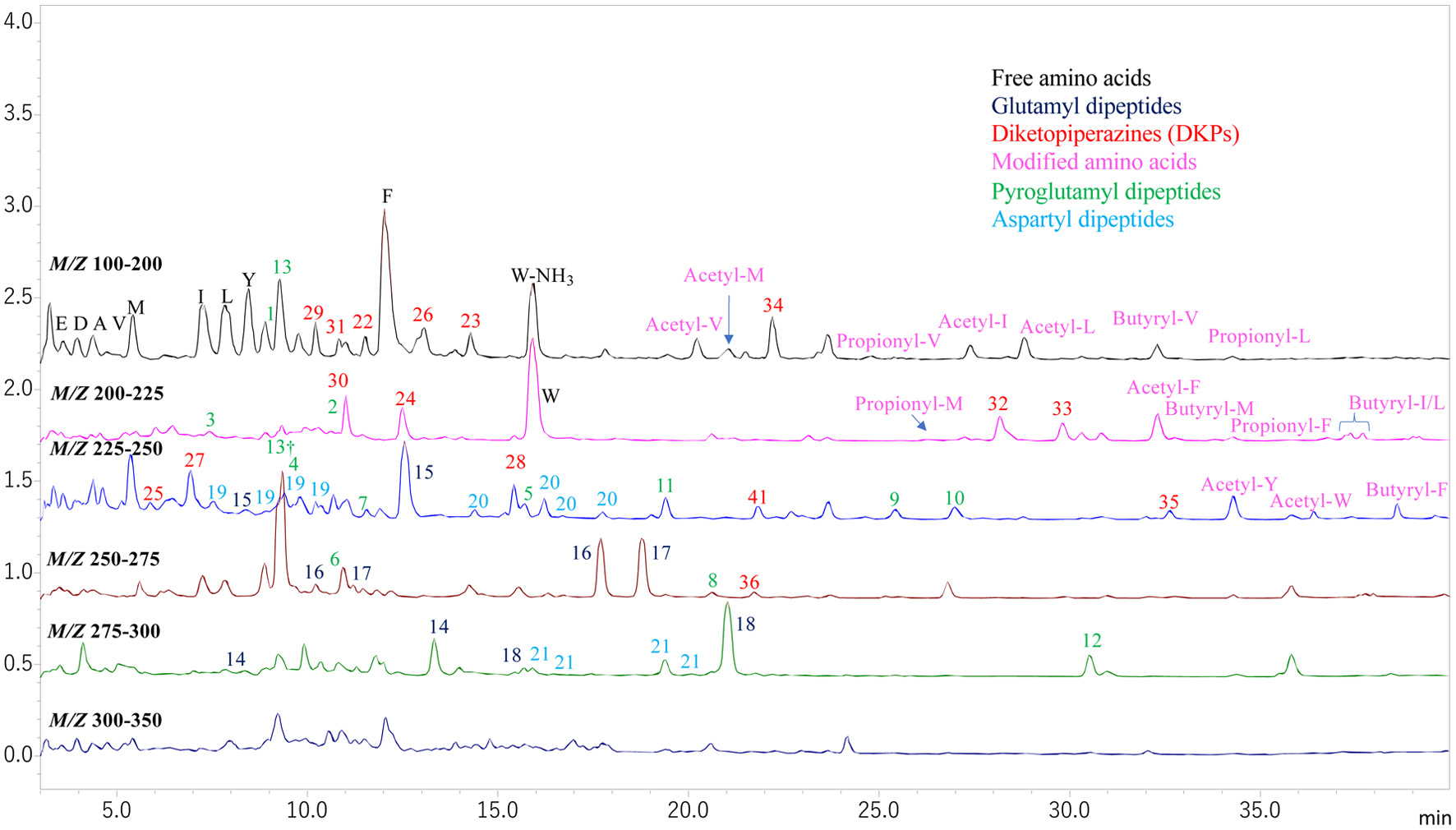
| Gly-Pro | 17.81 | 343 | 172 | 30 (Gly*), 70.0 (Pro*), 116.0 (y1), 171.0 (AccQ, b1), 173.3 (y2), 227.8 (b2) & 343.3 |
| Glu-Pro | 19.50 | 415 | 244 | 84 (Glub), 116 (y1), 171.0 (AccQ, b1), 227 (z2), 272 (a2), 272 (x2), 300 (b2) & 415 |
| Ala-Pro | 20.35 | 357 | 186 | 44.2 (Ala*), 70.0 (Pro*), 116.1 (y1), 171.0 (AccQ, b1), 186.8 (y2), 242.2 (b2) & 357.0 |
| Ser-Pro | 17.65 | 373 | 202 | 60 (Ser*), 70 (Pro*), 116 (y2), 171 (AccQ, b1), 258 (b2), 203 (y2) & 373 |
| Pro-Pro | 20.46 | 383 | 212 | 70.4 (Pro*), 116.1 (y1), 171.1 (AccQ, b1), 212.8 (y2), 240 (a2), 240 (x2), 267.9 (b2) & 383.0 |
| Val-Pro | 26.22 | 385 | 214 | 72.3 (Val*), 70.2 (Pro*), 116.2 (y1), 171.1 (AccQ, b1), 215.1 (y2), 242.1 (a2), 269.9 (b2) & 385.0 |
| Ile-Pro | 29.95 | 399 | 228 | 70.2 (Pro*), 86.0 (Ile*), 116.4 (y1), 171.1 (AccQ, b1), 228.8 (y2), 256.3 (x2), 284.1 (b2) & 399.0 |
| Leu-Pro | 30.47 | 399 | 228 | 70.6 (Pro*), 86.2 (Leu*), 116.4 (y1), 171.1 (AccQ, b1), 228.8 (y2), 256.3 (x2), 284.1 (b2) & 399.0 |
| Glu-Pro | 19.63 | 415 | 244 | 70.4 (Pro*), 84 (Glub) 102.2 (Glu*), 116.0 (y1), 171.1 (AccQ, b1) 187 (c1), 272 (a2), 300 (b2) & 415 |
| Gly-Pro-Ala | 17.58 | 414 | 243 | 44 (Gly*), 127, 155, 171.1 (AccQ, b1), 187,200, 228, 244 & 414 |
| Ser-Pro-Asp | 19.00 | 488 | 317 | 60 (Ser*), 171.1 (AccQ, b1), 255 (z2), 318 (y3), 355 (b3) & 488 |
| Gly-Pro-Thr | 20.35 | 444 | 273 | 70 (Pro*), 74 (Thr*) 114, 127, 145, 171 (AccQ, b1), 200 (a2), 217 (y2), 228 (b2) & 444 |
| Ala-Hyp | 16.26 | 373 | 202 | 44 (Ala*), 86,3 (Hyp*), 132.1 (y1), 171.0 ( AccQ, b1), 203.0, (y2), 242.3 (b2) & 373.0 |
| Ile-/Hyp | 25.56 | 415 | 244 | 86.0 (Ile*), 86.0 (Hyp*), 132.4 (y1), 171.1 (AccQ, b1), 245.1 (y2), 256.1 (a2), 284.1(b2) & 415.0 |
| Leu/Hyp | 26.00 | 415 | 244 | 86.0 (Ile*), 86.0 (Hyp*), 132.4 (y1), 171.1 (AccQ, b1), 245.1 (y2), 256.1 (a2), 284.1(b2) & 415.0 |
| Phe-Hyp | 21.59 | 449 | 278 | 145, 171 (AccQ, b1), 279 (y2), 318 (b2) & 449 |
| Gly-Pro-Hyp | 16.80 | 456 | 285 | 70 (Pro*), 86 (Hyp*), 127 (a3), 132 (y1), 171 (AccQ, b1), 229 (y2), 286 (y3) & 456 |
| Gly-Ile | 23.00 | 359 | 188 | 30 (Gly*), 86 (Ile*), 128, 132 (y1), 145, 171 (AccQ, b1), 189 (y2), 200 (a2), 228 (b2), & 359 |
| Gly-Leu | 23.50 | 359 | 188 | 30 (Gly*), 86 (Ile*), 128, 132 (y1), 145, 171 (AccQ, b1), 189 (y2), 200 (a2), 228 (b2), & 359 |
| Gly-Ile (Heavy type) | 23.00 | 360 | 189 | 86/87 (Ile*), 171/172 (AccQ, b1), 190 (y2), 201 (a2), 229 (b2), & 359 |
| Gly-Leu (Heavy type) | 23.50 | 360 | 189 | 86/87 (Leu*), 171/172 (AccQ, b1), 190 (y2), 201 (a2), 229 (b2), & 359 |
| Gly-His | 10.14 | 383 | 212 | 110 (His*), 156 (y1), 171 (AccQ, b1), 200 (a1), 213 (y2), 338 (a3), 365 (b3) & 383 |
| Pro-Glu | 13.75 | 415 | 244 | 70 (Pro*), 102 (Glu*), 171.1 (AccQ, b1), 264.1, 244.2, 367.4 & 415 |
| Gln-Pro | 13.50 | 414 | 243 | 84 (Glnb), 116 (y1), 171 (AccQ, b1), 244 (y2) & 414 |
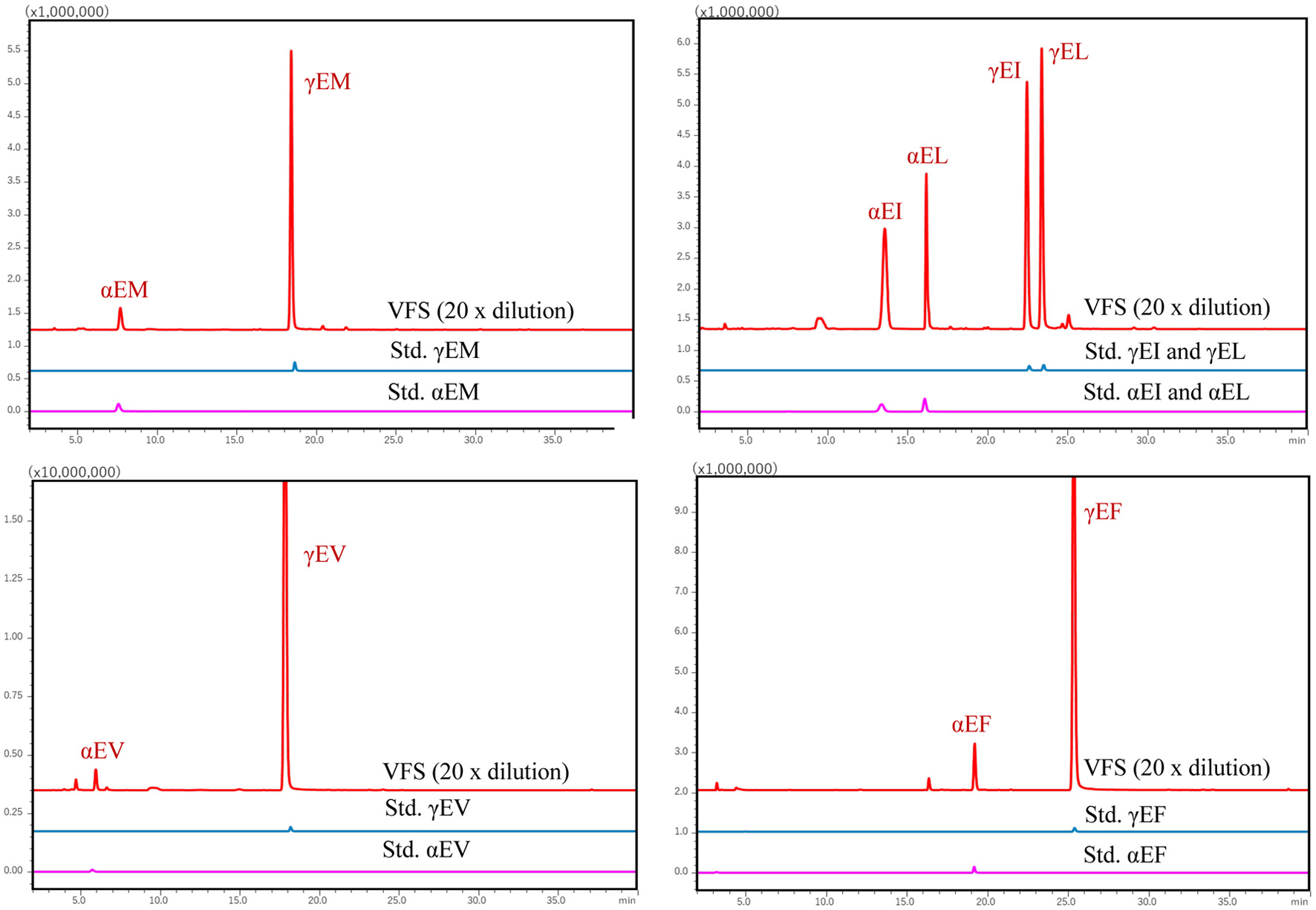
| Glu-Met | 21.00 | 449 | 278 | 84 (Glub), 171 (AccQ, b1), 272 (a2), 249 (x2), 279 (y2), 300 (b2) & 449 |
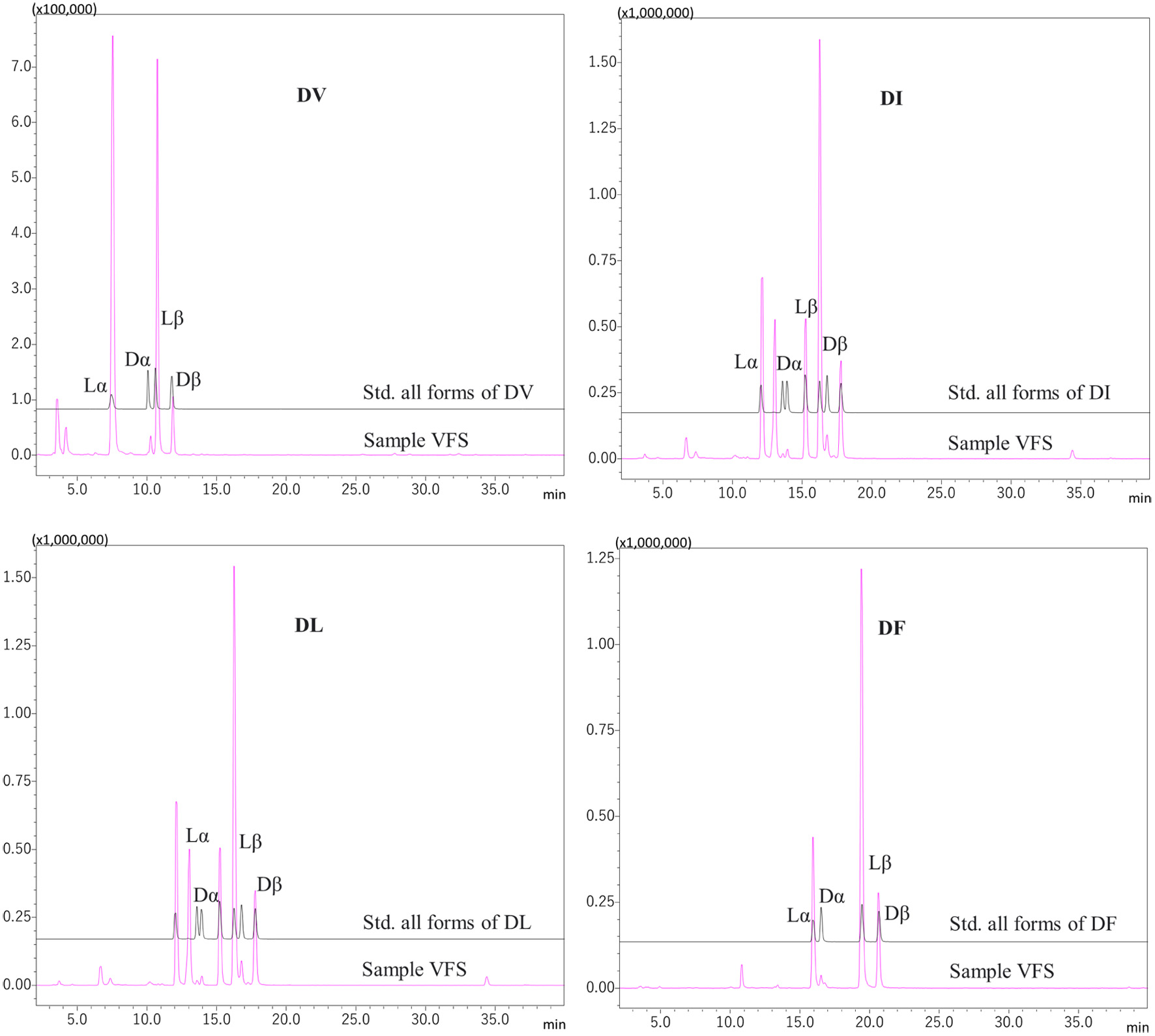
| Asp-Ile | 21.50 | 417 | 246 | 88 (Asp*), 132 (y1), 171 (AccQ, b1), 247 (y2), 286 (b2) & 417 |
| Asp-Leu | 22.00 | 417 | 246 | 88 (Asp*), 132 (y1), 171 (AccQ, b1), 247 (y2), 286 (b2) & 417 |
| Ala-Ile | 24.68 | 373 | 202 | 44 (Ala*), 86 (Ile*), 171 (AccQ, b1), 214 (a2), 242 (b2) & 373 |
| Ala-Leu | 24.78 | 373 | 202 | 44 (Ala*), 86 (Leu*), 171 (AccQ, b1), 214 (a2), 242 (b2) & 373 |
| Asn-Glu | 15.20 | 432 | 261 | 87 (Asn*), 70 (Asnb), 171 (AccQ, b1), 258 (a2), 286 (b2) & 433 |
| Val-Gly | 20.30 | 345 | 174 | 72 (Val*), 55 (Valb), 171 (AccQ, b1), 242 (a2), 270 (b2) & 345 |
| Ala-Val | 12.10 | 359 | 188 | 44 (Ala*), 171 (AccQ, b1), 189 (y2), 215 (a2) & 359 |
| Gly-Gly-Gly | 11.10 | 360 | 189 | 171 (AccQ, b1), 190 (y3), 200 (a2) & 360 |
| Gly-Met | 19.70 | 377 | 206 | 171 (AccQ, b1),186 (c1), 207 (y2), 228 (b2) & 377 |
| Ala-Glu | 16.40 | 389 | 218 | 44 (Ala*), 171 (AccQ, b1), 148 (y1), 219 (y2), 243 (b2) & 389 |
| Gly-Phe | 20.35 | 393 | 222 | 30 (Gly*), 120 (Phe*), 166 (y1), 171 (AccQ, b1), 201 (a2), 228 (b2) & 393 |
| Gly-Gly-Pro | 15.90 | 400 | 229 | 30 (Gly*), 70 (Pro*), 116 (y1), 171 (AccQ, b1), 200 (a2), 173 (y2), 227 (b2) & 400 |
| Glu-Ile | 25.00 | 431 | 260 | 84 (Glub), 86 (Ile*), 102 (Glu*), 132 (y1), 171 (AccQ, b1), 261 (y2), 300 (b2) & 431 |
| Glu-Leu | 26.00 | 431 | 260 | 84 (Glub), 86 (Leu*), 102 (Glu*), 132 (y1), 171 (AccQ, b1), 261 (y2), 300 (b2) & 431 |
| Glu-Phe | 27.00 | 465 | 294 | 120 (Phe*), 171 (AccQ, b1), 278 (z2), 278 (b2), 295 (y2) & 465 |
| Glu-Val | 21.50 | 465 | 294 | 72 (Val*), 102 (Glu*) 171 (AccQ, b1), 229 (z2) & 417 |
| Ala-Glu-Asn | 25.50 | 431 | 260 | 44 (Ala*), 171 (AccQ, b1), 261 (b3) & 431 |
| Glu-Gly | 14.52 | 375 | 204 | 102 (Glu*), 171 (AccQ, b1), 272 (a2) & 375 |