Figures
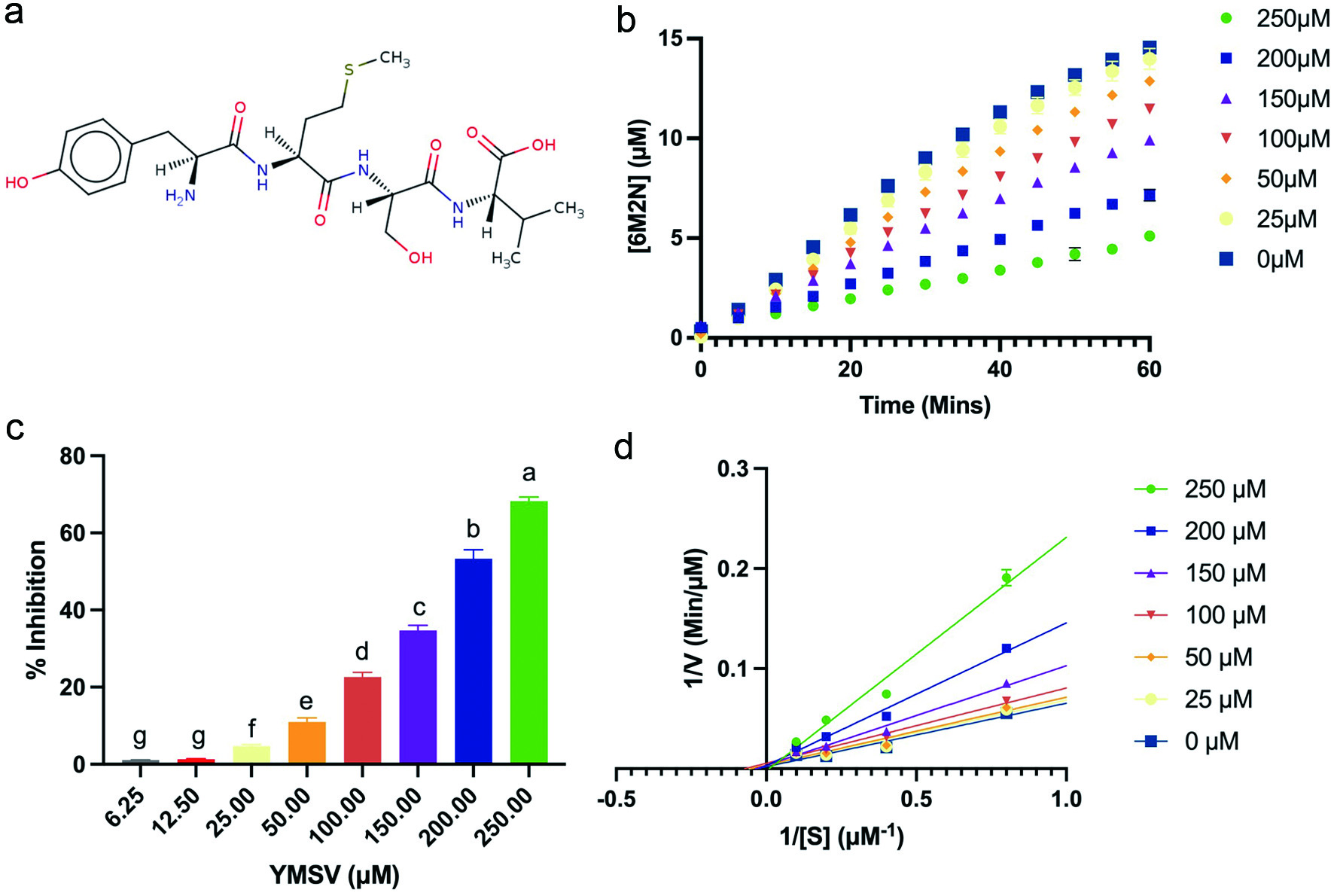
Figure 1. Effect of YMSV on the activity of soluble epoxide hydrolase (sEH). (a) Structure of peptide YMSV; (b) plots of the product (6M2N) formed at constant [S] (5 µM PHOME) over time at different concentrations of the inhibitor, YMSV; (c) concentration-dependent sEH inhibitory activity of YMSV; (d) Lineweaver-Burk double reciprocal plot of sEH at different YMSV concentration.
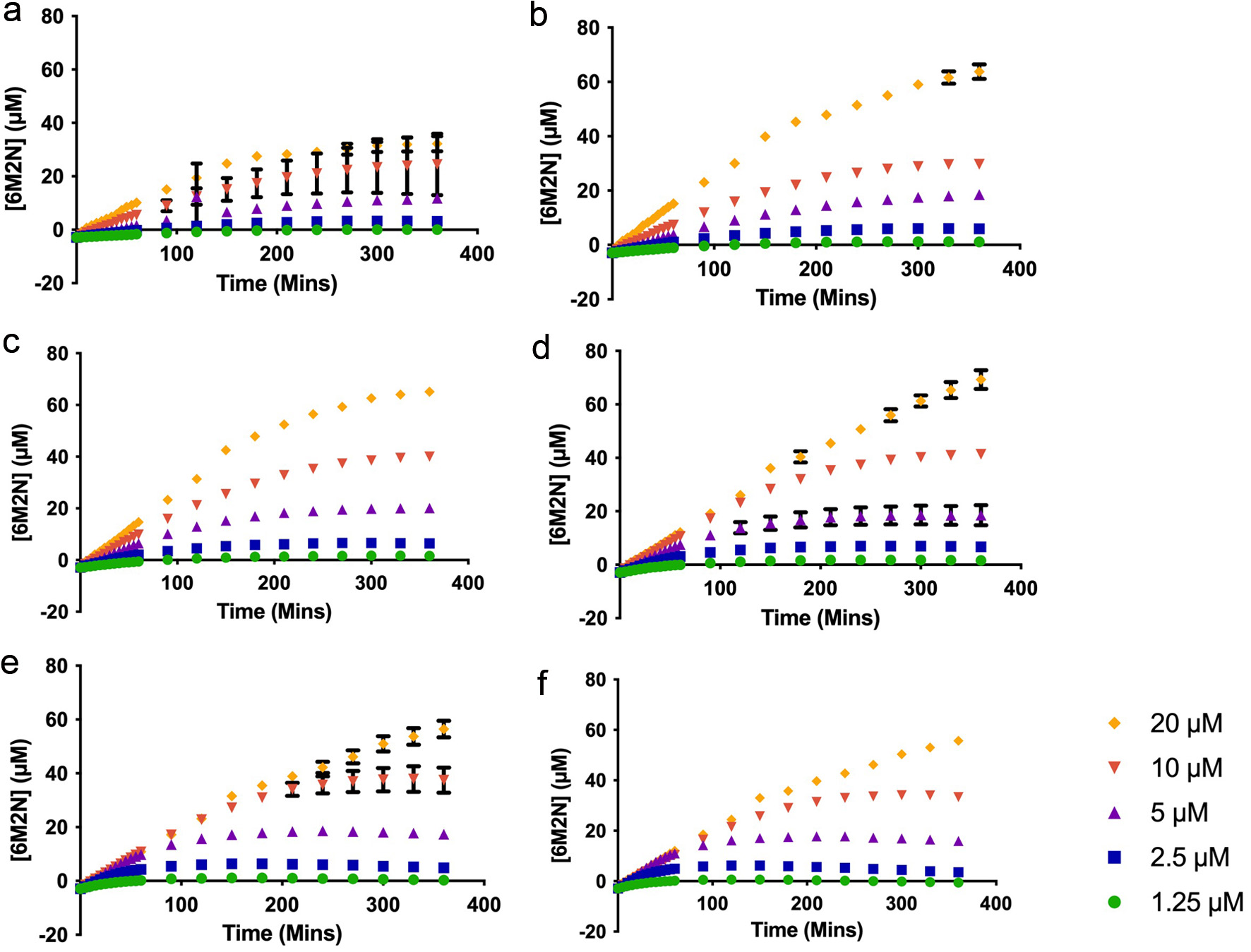
Figure 2. Plots of the product (6M2N) formed by human soluble epoxide hydrolase at the substrate (PHOME) concentrations of 1.25–20 µM in the presence of (a) 250 µM (b) 200 µM (c) 150 µM (d) 100 µM (e) 50 µM (f) 25 µM inhibitor (YMSV).
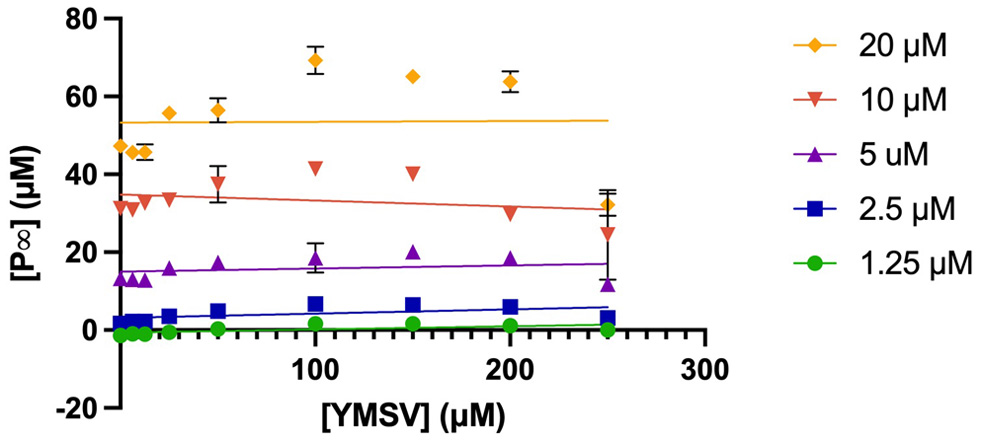
Figure 3. Plots of P∞ against varying concentrations of the inhibitor, YMSV with changing [S] (PHOME = 1.25−20 µM).
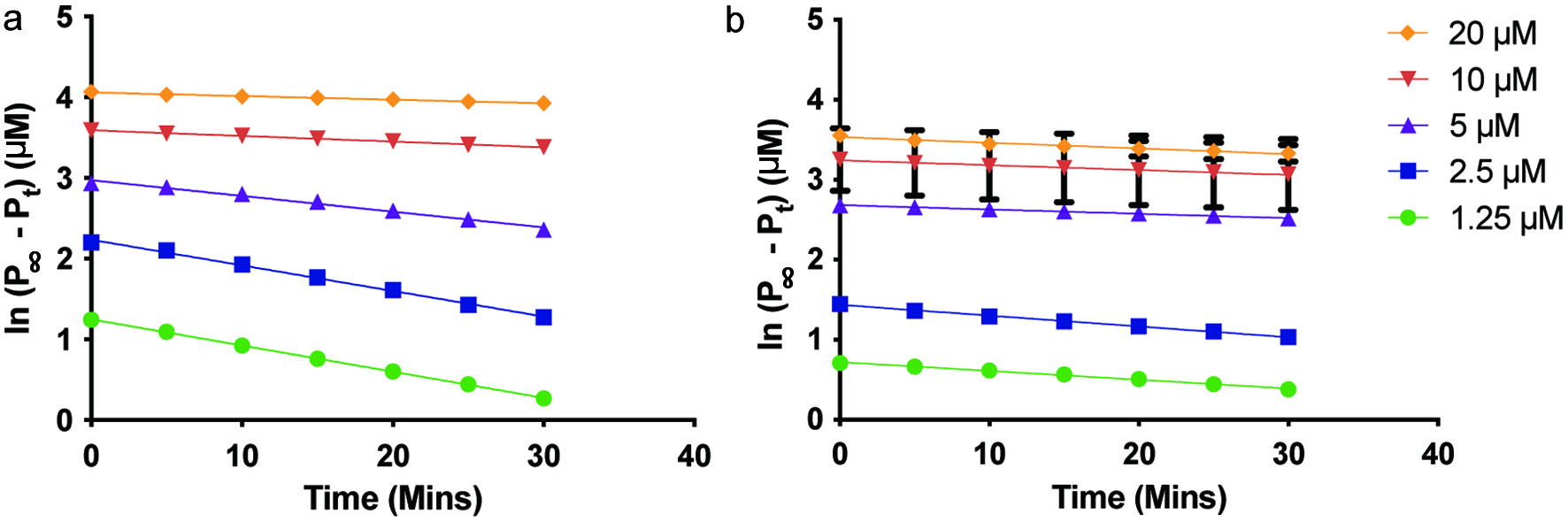
Figure 4. Plots of ln([P]∞ − [P]) vs. time to determine the apparent inactivation rate constant of soluble epoxide hydrolase in the presence of (a) 25 µM YMSV, and (b) 250 µM YMSV.
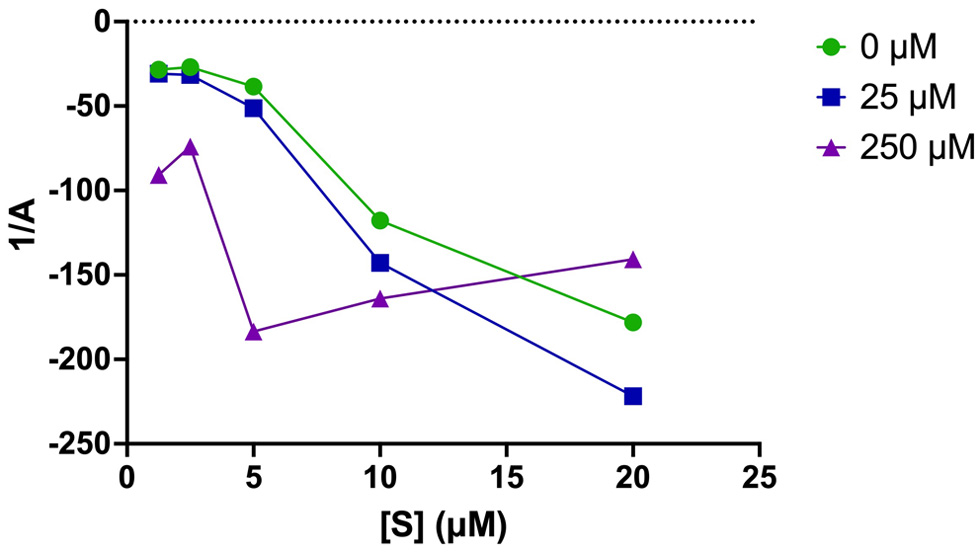
Figure 5. Plots of the reciprocal of apparent inactivation rate constant (A) vs. [S] in the absence and presence of different concentrations of inhibitor (25 and 250 µM YMSV).
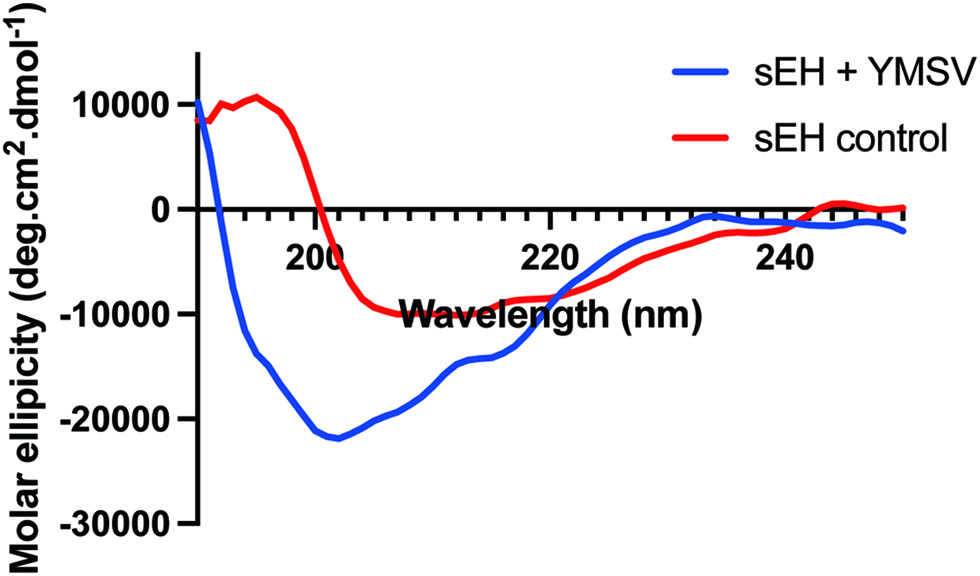
Figure 6. Circular dichroism spectra of soluble epoxide hydrolase in the absence (sEH control) and presence (sEH+YMSV) of the inhibitor peptide.
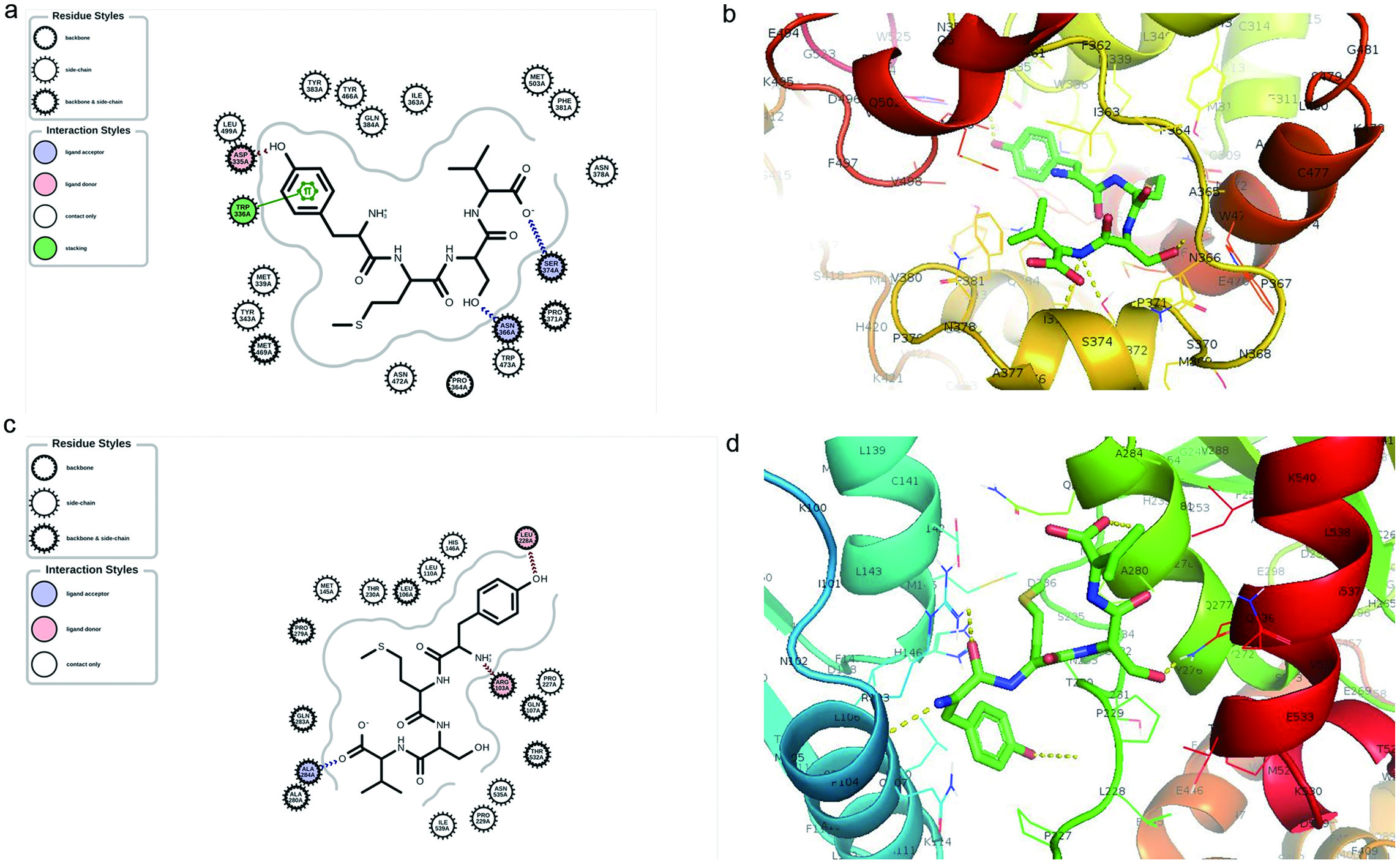
Figure 7. Molecular docking simulation showing (a) 2D and (b) 3D binding interaction of YMSV peptide and the active site of human soluble epoxide hydrolase (sEH), (c) 2D and (d) 3D binding interaction of YMSV peptide and human sEH upon blind docking.
Tables
Table 1. Kinetics parameters of sEH catalyzed reaction in the absence and presence of different concentrations of the YMSV peptide
| YMSV (µM) | Vmax (μM/min) | Km (μM) |
|---|
| Km, Michaelis constant in the absence and presence of the inhibitor; Vmax, maximum reaction velocity in the absence and presence of the inhibitor; ns, not significant (p ≥ 0.05); *, significant at 0.01 > p > 0.05); **, significant at 0.01 > p > 0.001; ***, significant at 0.001 > p > 0.0001; ****, significant at p < 0.0001 compared to control sEH (at 0 µM inhibitor). |
| 0 | 110.30 ± 0.36 | 3.514 ± 0.02 |
| 25 | 89.31 ± 4.55*** | 2.928 ± 0.11ns |
| 50 | 90.06 ± 1.29*** | 3.216 ± 0.05ns |
| 100 | 91.42 ± 1.54*** | 4.648 ± 0.17ns |
| 150 | 103.60 ± 0.90ns | 7.375 ± 0.14** |
| 200 | 98.61 ± 0.66* | 11.037 ± 0.16**** |
| 250 | 128.43 ± 9.92*** | 24.830 ± 2.80**** |
Table 2. In silico absorption, distribution, metabolism, and excretion (ADME) profile of the YMSV peptide and AUDA generated using SwissADME
| YMSV | AUDA |
|---|
| Abbreviations: Molecular Weight (MW) (g/mol); Number Of Rotatable Bonds (ROTB); Hydrogen Bond Donors (HBD); Hydrogen Bond Acceptors (HBA); Estimated Solubility (ESOL) (Delaney, 2004); Toxin Support Vector Machine (SVM) score (BIOPEP and ToxinPred) (Gupta et al., 2013); Topological Polar Surface Area (TPSA); Logarithm Of Compound Partition Coefficient Between N-Octanol And Water (ClogP); Bioavailability score, probability of F > 10% in rat (Martin, 2005); Lipinski Filter (Lipinski’s Rule-Of-5); Gastrointestinal Absorption (GIA); P-Glycoprotein Substrate, And Cytochrome P450 3A4 (CYP3A4) Inhibitor; Blood Brain Barrier (BBB) permeation. |
| Physicochemical properties | MW (g/mol) | 498.59 | 392.58 |
| ROTB (n) | 17 | 15 |
| HBA (n) | 8 | 3 |
| HBD (n) | 7 | 3 |
| ESOL (Log S) | 0.23 | - 4.98 |
| Lipophilicity | TPSA (Å2) | 216.38 | 78.43 |
| CLogP (o/w) | −0.39 | 4.69 |
| Drug-likeness and pharmacokinetics | Bioavailability score | 0.17 | 0.56 |
| GIA | Low | High |
|---|
| Abbreviations: Molecular Weight (MW) (g/mol); Number Of Rotatable Bonds (ROTB); Hydrogen Bond Donors (HBD); Hydrogen Bond Acceptors (HBA); Estimated Solubility (ESOL) (Delaney, 2004); Toxin Support Vector Machine (SVM) score (BIOPEP and ToxinPred) (Gupta et al., 2013); Topological Polar Surface Area (TPSA); Logarithm Of Compound Partition Coefficient Between N-Octanol And Water (ClogP); Bioavailability score, probability of F > 10% in rat (Martin, 2005); Lipinski Filter (Lipinski’s Rule-Of-5); Gastrointestinal Absorption (GIA); P-Glycoprotein Substrate, And Cytochrome P450 3A4 (CYP3A4) Inhibitor; Blood Brain Barrier (BBB) permeation. |
| Lipinski filter | No | Yes |
| P-glycoprotein substrate | Yes | No |
| CYP 3A4 inhibitor | No | Yes |
| BBB permeation | No | No |






