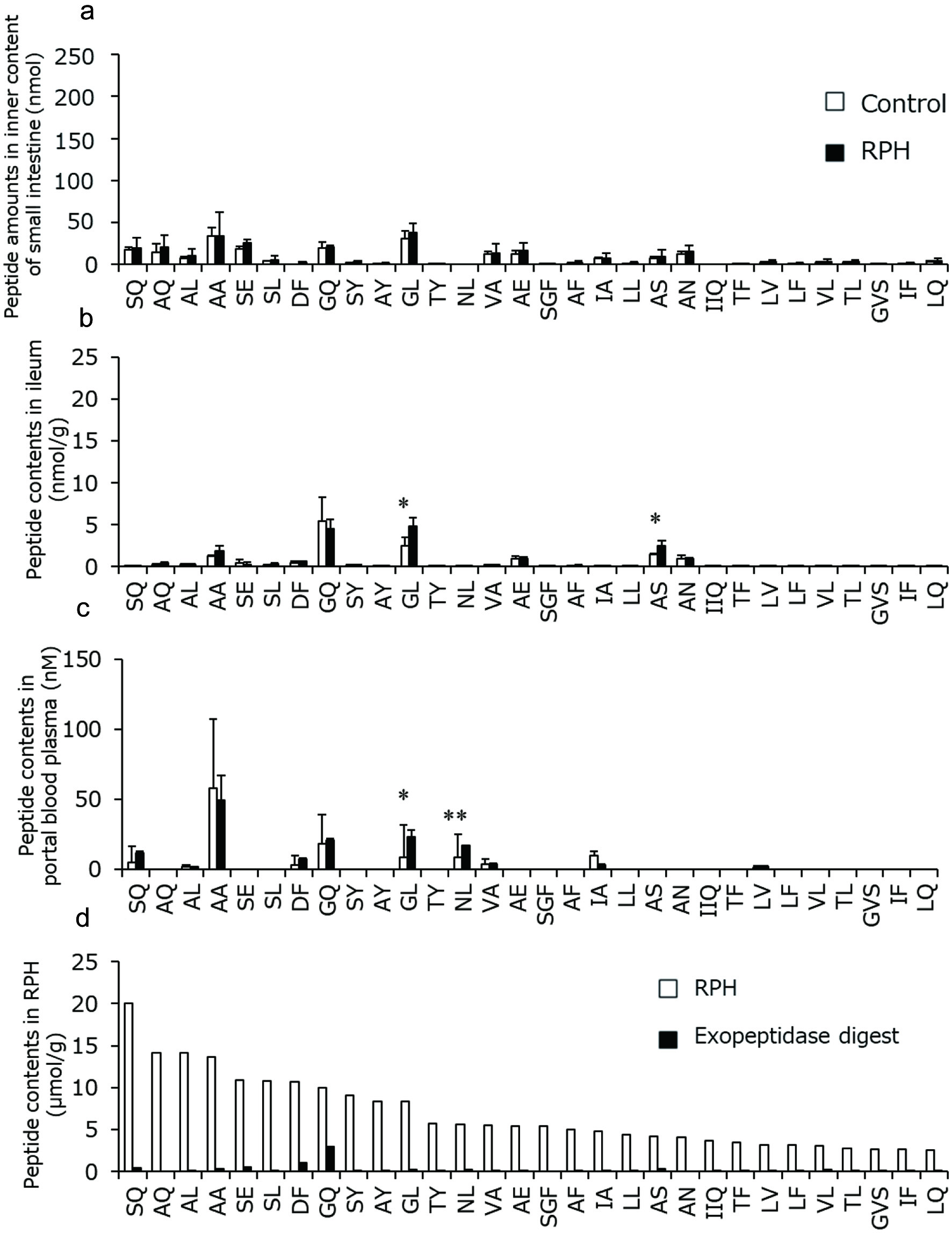
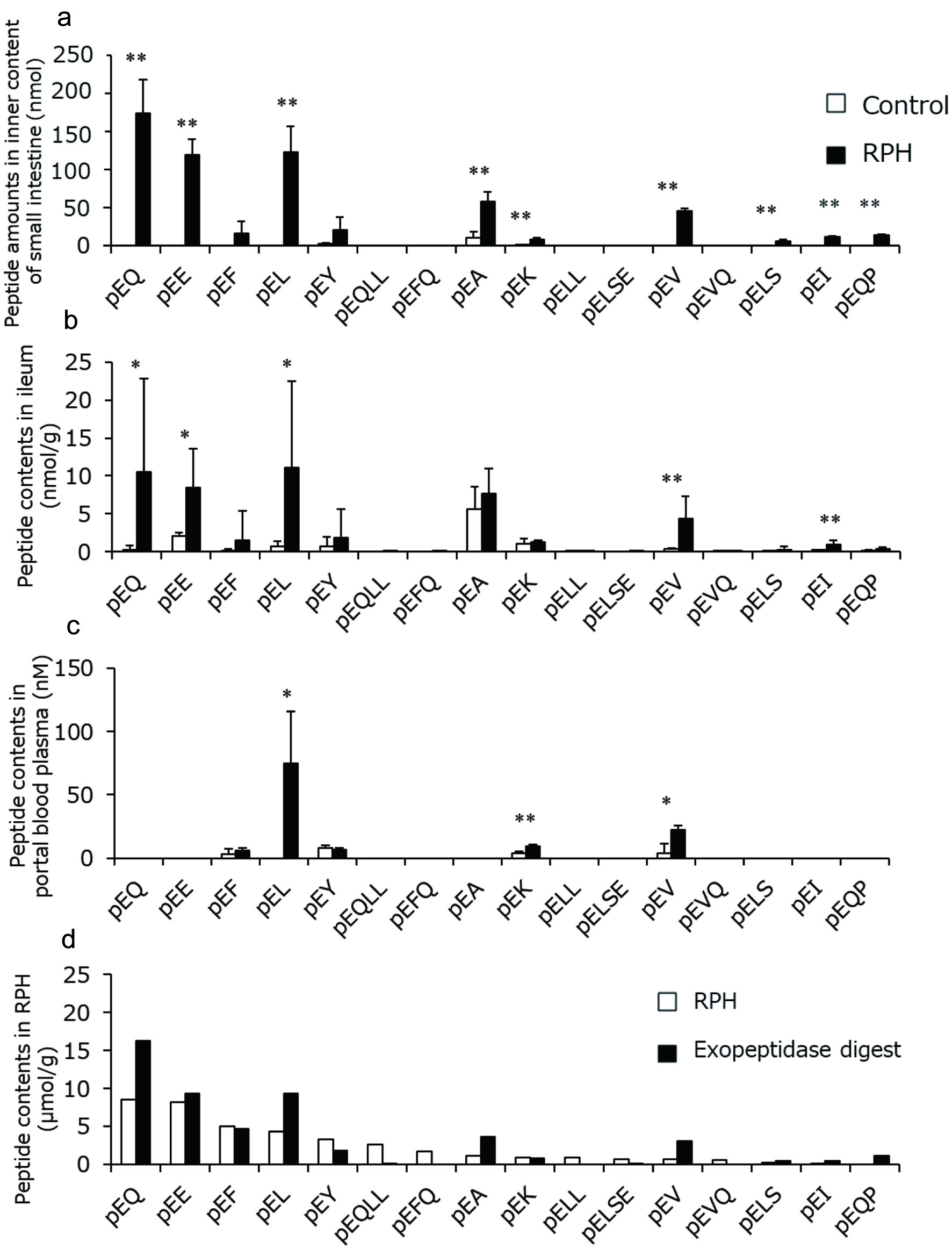
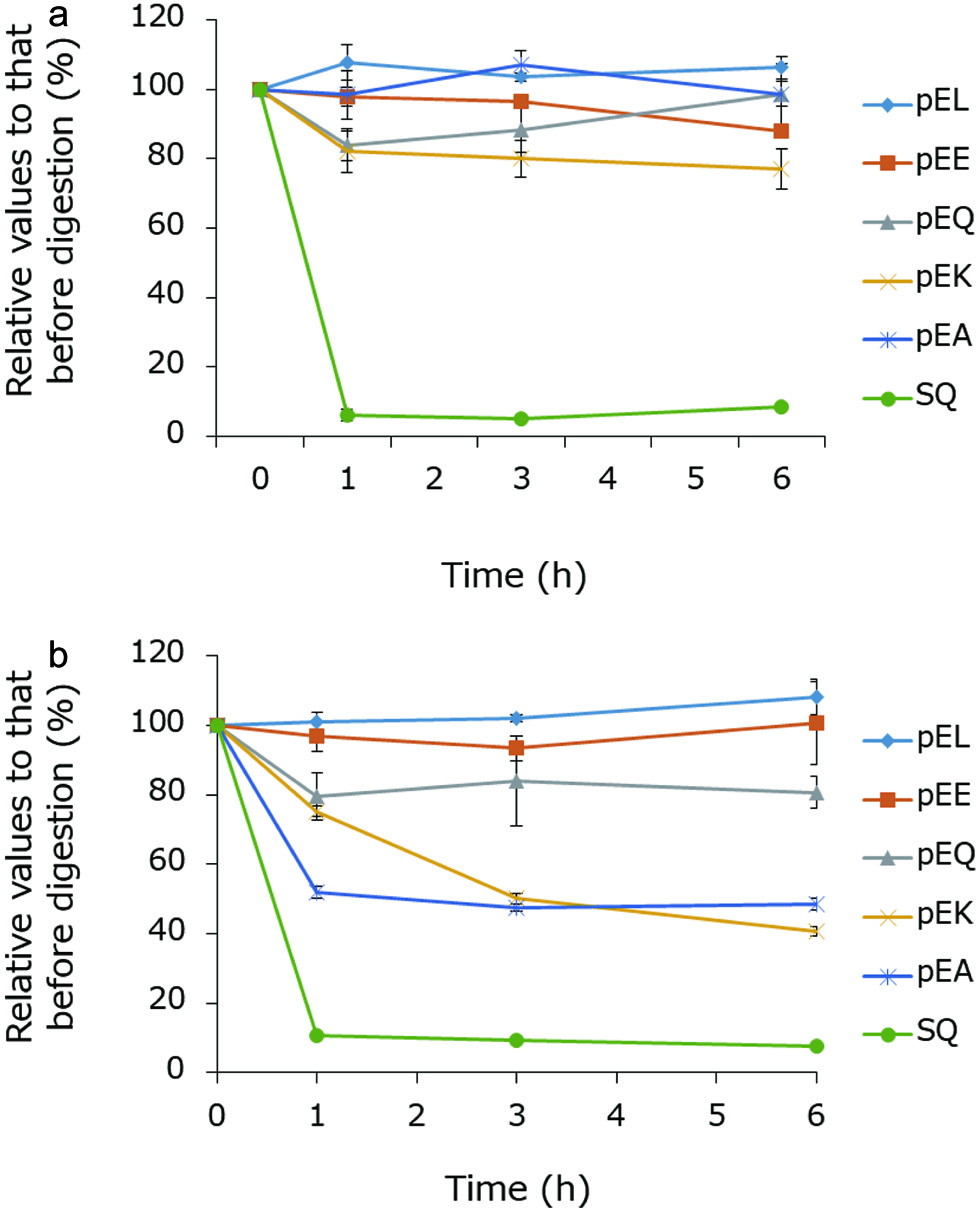
| 1 | 36 | AS | 347 | 170.9 (AccQ, b1), 176.9 (y1), 213.9 (a2), 242.0 (b2) |
| 2 | 36 | SA | 347 | 60.5 (Ser*), 171.0 (AccQ, b1), 177.2 (y2), 257.8 (b2) |
| 3 | 36, 37 | AA | 331 | 161.0 (y2), 171.0 (AccQ, b1), 241.8 (b2) |
| 4 | 36 | GQ | 374 | 84.1 (Gln**), 146.7 (y1), 170.9 (AccQ, b1), 203.7 (y2), 227.8 (b2) |
| 5 | 36 | AN | 374 | 44.4 (Ala*), 132.9 (y1), 170.9 (AccQ, b1), 203.9 (y2), 214.1 (a2), 241.7 (b2) |
| 6 | 36 | AQ | 388 | 44.5 (Ala*), 147.0 (y1), 170.9 (AccQ, b1), 213.9 (y2), 217.9 (y2), 241.9 (b2) |
| 7 | 36 | AT | 361 | 44.0 (Ala*), 119.7 (y1), 171.0 (AccQ, b1), 190.8 (y2), 213.7 (a2), 242.1 (b2) |
| 8 | 36 | AE | 389 | 148.1 (y1), 170.9 (AccQ, b1), 213.9 (a2), 219.0 (y2), 242.1 (b2) |
| 9 | 36 | TA | 361 | 170.9 (AccQ, b1), 190.7 (y2), 272.1 (b2) |
| 10 | 36 | PA | 357 | 70.1 (Pro*), 171.0 (AccQ, b1), 186.9 (y2), 240.0 (a2) |
| 11 | 36 | VS | 375 | 72.3 (Val*), 171.0 (AccQ, b1), 205.0 (y2), 242.0 (a2), 270.2 (b2) |
| 12 | 36 | VT | 389 | 72.2 (Val*), 170.9 (AccQ, b1), 218.8 (y2), 241.9 (a2), 269.8 (b2) |
| 13 | 36 | VA | 359 | 72.2 (Val*), 170.9 (AccQ, b1), 189.0 (y2), 242.0 (a2), 269.9 (b2) |
| 14 | 36 | IS, LS1 | 389 | 86.1 (Ile/Leu*), 171.0 (AccQ, b1), 219.0 (y2), 256.0 (a2), 283.6 (b2) |
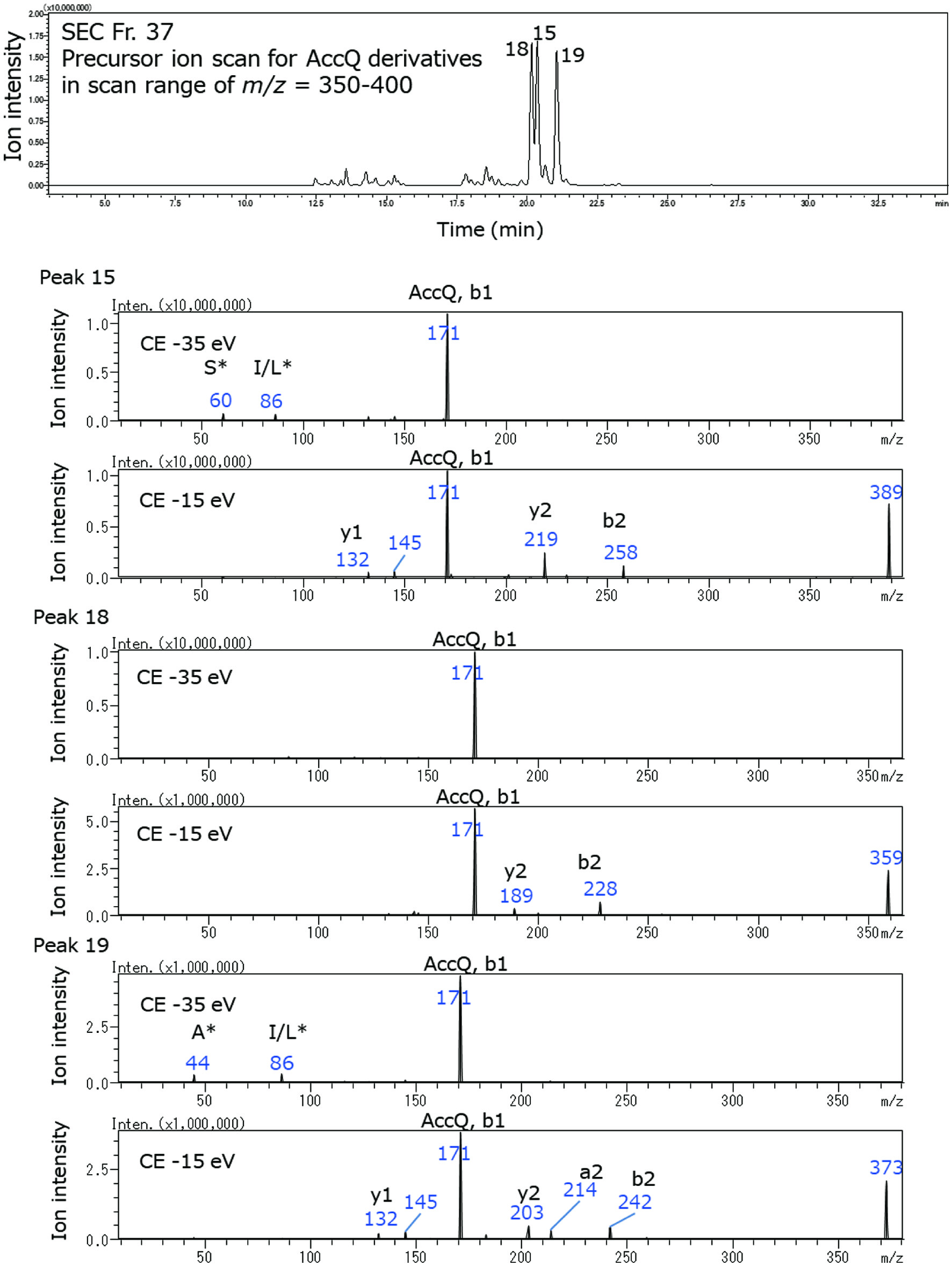
| 15 | 36, 37 | SI, SL1 | 389 | 171.0 (AccQ, b1), 219.0 (y2), 258.0 (b2) |
| 16 | 36 | IA, LA1 | 373 | 86.1 (Ile/Leu*), 170.9 (AccQ, b1), 203.0 (y2), 255.9 (a2), 283.9 (b2) |
| 17 | 36 | VV | 387 | 72.4 (Val*), 170.8 (AccQ, b1), 217.0 (y2), 242.1 (a2), 270.3 (b2) |
| 18 | 37 | GI, GL1 | 359 | 170.9 (AccQ, b1), 188.9 (y2), 227.9 (b2) |
| 19 | 37 | AI, AL1 | 373 | 132.1 (y1), 170.9 (AccQ, b1), 203.1 (y2), 213.9 (a2), 241.9 (b2) |
| 20 | 42–45 | GF | 393 | 171.0 (AccQ, b1), 227.9 (b2) |
| 21 | 36, 37 | SQ | 404 | 60.2 (Ser*), 83.6 (Gln**), 147.0 (y1), 170.9 (AccQ, b1), 233.8 (y2), 230.0 (a2), 257.8 (b2) |
| 22 | 36 | DT2 | 405 | 74.2 (Thr*), 171.1 (AccQ, b1), 234.7 (y2), 258.2 (a2) |
| 23 | 36 | EQ2 | 446 | 83.8 (Glu**, Gln**), 147.2 (y1), 171.1 (AccQ, b1), 272.3 (a2), 275.5 (y2), 299.6 (b2) |
| 24 | 36, 37 | TE | 419 | 74.4 (Thr*), 84.4 (Glu**), 147.7 (y1), 170.8 (AccQ, b1), 243.7 (a2), 248.7 (y2) |
| 25 | 36 | GVS | 432 | 72.1 (Val*), 106.1 (y1), 170.9 (AccQ, b1), 199.9 (a2), 228.1 (b2), 327.1 (b3), 385.6 (a4) |
| 26 | 36 | GVA | 416 | 72.4 (Val*), 90.1 (y1), 170.9 (AccQ, b1), 188.8 (y2), 200.0 (a2), 228.1 (b2), 327.0 (b3) |
| 27 | 36 | VQ | 416 | 71.9 (Val*), 101.1 (Gln*), 147.1 (y1), 171.0 (AccQ, b1), 242.1 (a2), 245.8 (y2), 269.8 (b2) |
| 28 | 36 | GIS, GLS2 | 446 | 86.3 (Ile/Leu*), 106.1 (y1), 171.0 (AccQ, b1), 199.7 (a2), 228.0 (b2), 276.0 (y3), 340.6 (b3) |
| 29 | 36, 37 | VE | 417 | 72.0 (Val*), 148.2 (y1), 170.9 (AccQ, b1), 242.1 (a1), 246.9 (y2), 270.1 (b2) |
| 30 | 36 | GAI, GAL2 | 430 | 44.3 (Ala*), 86.3 (Ile/Leu*), 132.1 (y1), 171.2 (AccQ, b1), 228.1 (b2), 259.8 (y2) |
| 31 | 36 | IQ, LQ1 | 430 | 84.1 (Gln**), 86.2 (Ile/Leu*), 146.9 (y1), 170.8 (AccQ, b1), 256.1 (a2), 259.8 (y2), 283.9 (b2) |
| 32 | 36 | IT, LT2 | 403 | 86.2 (Ile/Leu*), 119.7 (y1), 171.0 (AccQ, b1), 232.5 (y2), 255.9 (a2) |
| 33 | 36, 37 | TI, TL1 | 403 | 86.1 (Ile/Leu*), 170.9 (AccQ, b1), 233.0 (y2), 272.0 (b2) |
| 34 | 36, 37 | IV, LV1 | 401 | 86.0 (Ile/Leu*), 171.1 (AccQ, b1), 230.9 (y2), 255.8 (a2), 284.0 (b2) |
| 35 | 36, 37 | VI, VL1 | 401 | 72.1 (Val*), 170.8 (AccQ, b1), 231.1 (y2), 242.3 (a2), 269.8 (b2) |
| 36 | 36, 37 | MI, ML1 | 433 | 86.2 (ile/Leu*), 104.2 (Met*), 171.0 (AccQ, b1), 274.0 (a2) |
| 37 | 36, 37 | II, IL, LI, LL1 | 415 | 86.2 (Ile/Leu*), 171.0 (AccQ, b1), 244.9 (y2), 284.1 (b2) |
| 38 | 37 | NE2 | 432 | 87.7 (Asn*), 101.7 (Glu*), 171.0 (AccQ, b1), 256.9 (a2), 261.9 (y2), 285.0 (b2) |
| 39 | 37 | SE | 405 | 60.4 (Ser*), 147.8 (y1), 171.0 (AccQ, b1), 234.9 (y2) |
| 40 | 37 | SM2 | 407 | 150.0 (y1), 170.9 (AccQ, b1), 236.9 (y2), 257.7 (b2) |
| 41 | 37 | NI, NL1 | 416 | 87.0 (Asn*), 132.0 (y1), 171.0 (AccQ, b1), 245.9 (y2), 256.8 (a2), 284.9 (b2) |
| 42 | 37 | DI, DL1 | 417 | 86.3 (Ile/Leu*), 131.7 (y1), 170.9 (AccQ, b1), 247.1 (y2), 286.0 (b2) |
| 43 | 37 | EI, EL1 | 431 | 84.1 (Glu**), 86.2 (Ile/Leu*), 132.4 (y1), 170.8 (AccQ, b1), 261.0 (y2), 271.9 (a2), 299.8 (b2) |
| 44 | 37 | IM, LM2 | 433 | 86.3 (Ile/Leu*), 170.8 (AccQ, b1), 256.1 (a2), 263.1 (y2), 284.3 (b2) |
| 45 | 41–43 | SF | 423 | 119.9 (Phe*), 166.0 (y1), 171.0 (AccQ, b1), 230.0 (a2), 253.0 (y2), 258.0 (b2) |
| 46 | 41, 42 | TF | 437 | 74.2 (Thr*), 166.0 (y1), 170.9 (AccQ, b1), 243.7 (a2), 267.0 (y2), 271.9 (b2) |
| 47 | 41–43 | AF | 407 | 120.1 (Phe*), 165.9 (y1), 171.0 (AccQ, b1), 213.8 (a2), 236.9 (y2), 241.9 (b2) |
| 48 | 41–43 | VF | 435 | 72.1(Val*), 170.9 (AccQ, b1), 242.0 (a2), 264.9 (y2), 270.0 (b2) |
| 49 | 42–45 | IF, LF1 | 449 | 86.2 (Ile/Leu*), 119.5 (Phe*), 171.0 (AccQ, b1), 256.1 (a2), 279.1 (y2), 283.8 (b2) |
| 50 | 43 | SY | 439 | 60.4 (Ser*), 136.1 (Tyr*), 171.0 (AccQ, b1), 182.1 (y1), 229.7 (a2), 257.9 (b2), 269.1 (y2) |
| 51 | 43 | AY | 423 | 171.0 (AccQ, b1), 182.0 (y1), 213.8 (a2), 241.9 (b2) |
| 52 | 36 | STS | 464 | 59.7 (Ser*), 106.1 (y1), 171.2 (AccQ, b1), 219.9 (a2), 257.6 (b2) |
| 53 | 36. 37 | GVE2 | 474 | 148.0 (y1), 170.8 (AccQ, b1), 227.9 (b2), 303.6 (y3) |
| 54 | 36, 37 | GIN, GLN2 | 473 | 85.8 (Ile/Leu*), 87.3 (Asn*), 133.1 (y1), 171.1 (AccQ, b1), 199.5 (a2), 227.8 (b2), 303.1 (y3) |
| 55 | 36 | GIT, GLT2 | 460 | 85.9 (Ile/Leu*), 119.8 (y1), 170.9 (AccQ, b1), 199.6 (a2), 228.1 (b2), 290.1 (y3), 340.6 (b3) |
| 56 | 36 | ASI, ASL1 | 460 | 44.1 (Ala*), 59.6 (Ser*), 85.7 (Ile/Leu*), 171.0 (AccQ, b1), 241.4 (b2), 289.5 (y3), 329.2 (b3) |
| 57 | 36 | AVI, AVL1 | 472 | 44.4 (Ala*), 71.9 (Val*), 170.4 (AccQ, b1), 214.1 (a2), 241.7 (b2), 301.7 (y3), 340.9 (b3) |
| 58 | 36 | IAI, IAL, LAI, LAL2 | 486 | 86.2 (Ile/Leu*), 170.7 (AccQ, b1), 283.7 (b2), 315.9 (y3), 355.3 (b3) |
| 59 | 37–41 | SGF | 480 | 60.6 (Ser*), 120.5 (Phe*), 165.9 (y1), 170.4 (AccQ, b1), 222.8 (y2), 258.2 (b2) |
| 60 | 37, 38 | GTF2 | 494 | 73.9 (Thr*), 119.7 (Phe*), 165.8 (y1), 227.7 (b2), 267.1 (y2), 300.7 (a3), 323.7 (y3) |
| 61 | 37–40 | GAF2 | 464 | 120.0 (Phe*), 166.0 (y1), 170.8 (AccQ, b1), 227.9 (b2) |
| 62 | 37–42 | FQ2 | 464 | 84.5 (Gln**), 119.9 (Phe*), 147.4 (y1), 170.7 (AccQ, b1), 289.6 (a2), 318.2 (b2) |
| 63 | 37 | AAF2 | 478 | 166.0 (y1), 170.8 (AccQ, b1), 241.8 (b2), 307.7 (y3) |
| 64 | 37 | GII, GIL, GLI, GLL2 | 472 | 86.0 (Ile/Leu*), 131.9 (y1), 170.7 (AccQ, b1), 200.2 (a2), 227.9 (b2), 340.8 (b3) |
| 65 | 41 | GGF2 | 450 | 120.1 (Phe*), 165.9 (y1), 171.1 (AccQ, b1), 199.7 (a2), 227.8 (b2), 279.8 (y3), 285.0 (b3) |
| 66 | 41 | DF | 451 | 88.2 (Asp*), 171.0 (AccQ, b1), 257.7 (a2), 280.7 (y2), 285.9 (b2) |
| 67 | 41 | FE2 | 465 | 119.9 (Phe*), 147.9 (y1), 171.0 (AccQ, b1), 294.6 (y2), 317.6 (b2) |
| 68 | 41, 42 | GVF2 | 492 | 72.0 (Val*), 119.4 (Phe*), 166.0 (y1), 171.0 (AccQ, b1), 199.8 (a2), 227.9 (b2), 326.5 (b3) |
| 69 | 42 | YQ2 | 480 | 83.8 (Gln**), 100.7 (Gln*), 136.0 (Tyr*), 147.0 (y1), 170.8 (AccQ, b1), 305.9 (a2), 333.6 (b2) |
| 70 | 42–44 | NF2 | 450 | 86.6 (Asn*), 165.8 (y1), 170.8 (AccQ, b1), 284.9 (b2) |
| 71 | 42–44 | EF2 | 465 | 84.0 (Glu**), 102.1 (Glu*), 120.2 (Phe*), 165.9 (y1), 171.0 (AccQ, b1), 299.8 (b2) |
| 72 | 43–45 | TY | 453 | 74.0 (Thr*), 136.0 (Tyr*), 271.8 (b2), 282.7 (y2) |
| 73 | 44, 45 | VY2 | 451 | 72.3 (Val*), 170.9 (AccQ, b1), 241.9 (a2), 269.8 (b2), 280.6 (y2) |
| 74 | 36 | STE2 | 506 | 59.9 (Ser*), 74.1 (Thr*), 147.6 (y1), 170.9 (AccQ, b1), 229.5 (a2), 257.7 (b2), 335.6 (y3) |
| 75 | 36 | LTE, ITE2 | 532 | 85.8 (Ile/Leu*), 147.7 (y1), 170.9 (AccQ, b1), 255.5 (a2), 361.6 (y3), 385.2 (b3) |
| 76 | 36 | TPI, TPL1 | 500 | 69.7 (Pro*), 85.7 (Ile/Leu*), 132.2 (y1), 170.5 (AccQ, b1), 228.8 (y2), 271.8 (b2), 330.1 (y3) |
| 77 | 36 | IIQ, ILQ, LIQ, LLQ1 | 543 | 86.1 (Ile/Leu*), 100.8 (Gln*), 147.1 (y1), 171.0 (AccQ, b1), 256.0 (a2), 284.0 (b2), 372.9 (y3), 396.9 (b3) |
| 78 | 37 | DVI, DVL1 | 516 | 85.9 (Ile/Leu*), 171.0 (AccQ, b1), 285.6 (b2), 384.9 (b3) |
| 79 | 37, 38 | IAF, LAF1 | 520 | 86.0 (Ile/Leu*), 165.6 (y1), 171.0 (AccQ, b1), 236.4 (y2), 256.1 (a2), 283.8 (b2), 354.7 (b3) |
| 80 | 41–44 | NIF, NLF1 | 563 | 86.1 (Ile/Leu*), 119.7 (Phe*), 170.8 (AccQ, b1), 257.1 (a2), 284.9 (b2), 397.8 (b3) |
| 81 | 41–44 | SIF, SLF1 | 536 | 60.2 (Ser*), 86.1 (Ile/Leu*), 119.9 (Phe*), 165.6 (y1), 170.9 (AccQ, b1), 229.7 (a2), 371.0 (b3) |