
Figure 1. Fragments of ovalbumin released by the action of the GI enzymes, chymotrypsin, trypsin, and pepsin.
| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Original Research
Volume 17, March 2022, pages 34-48
In silico investigation of molecular targets, pharmacokinetics, and biological activities of chicken egg ovalbumin protein hydrolysates
Figures

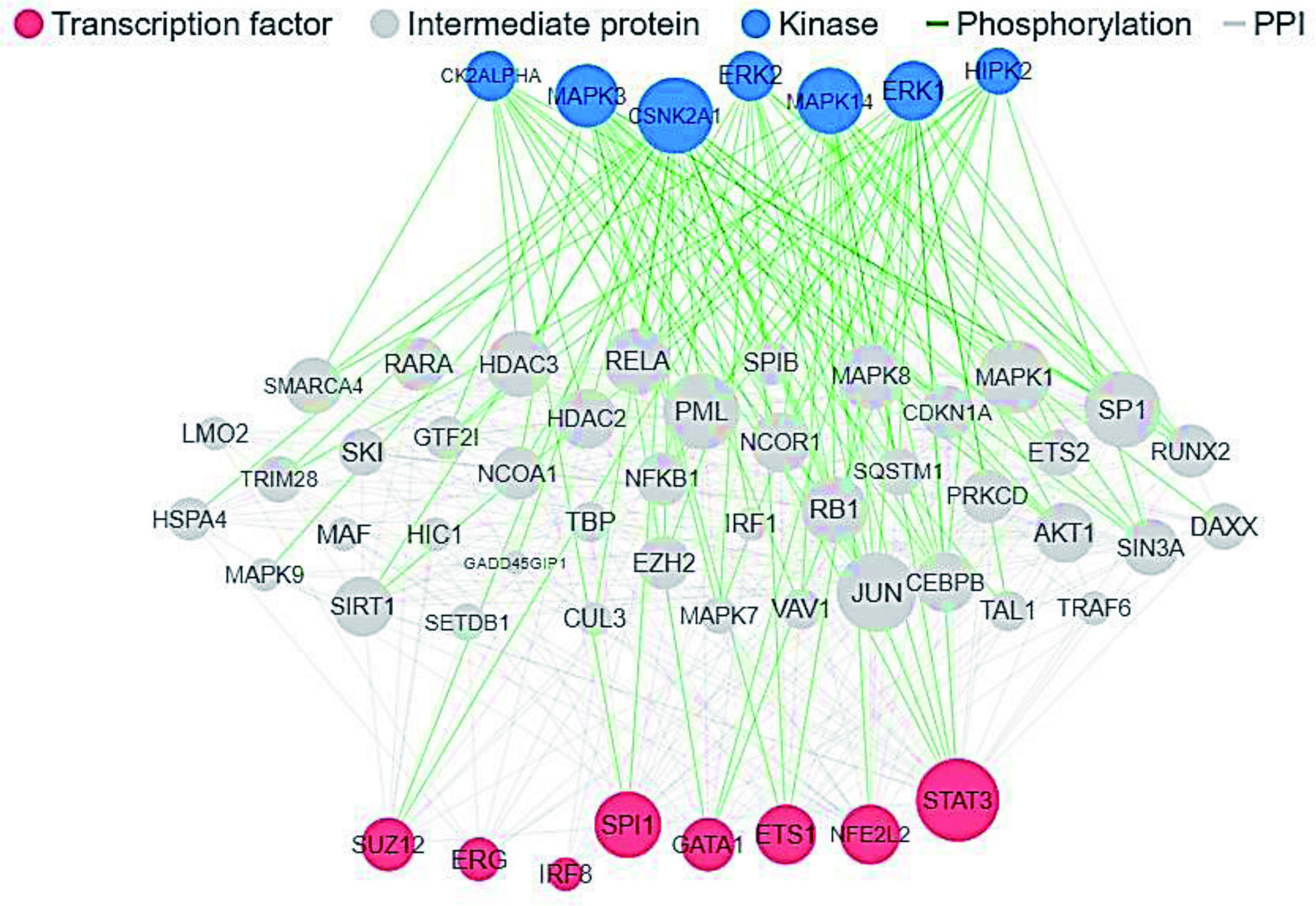
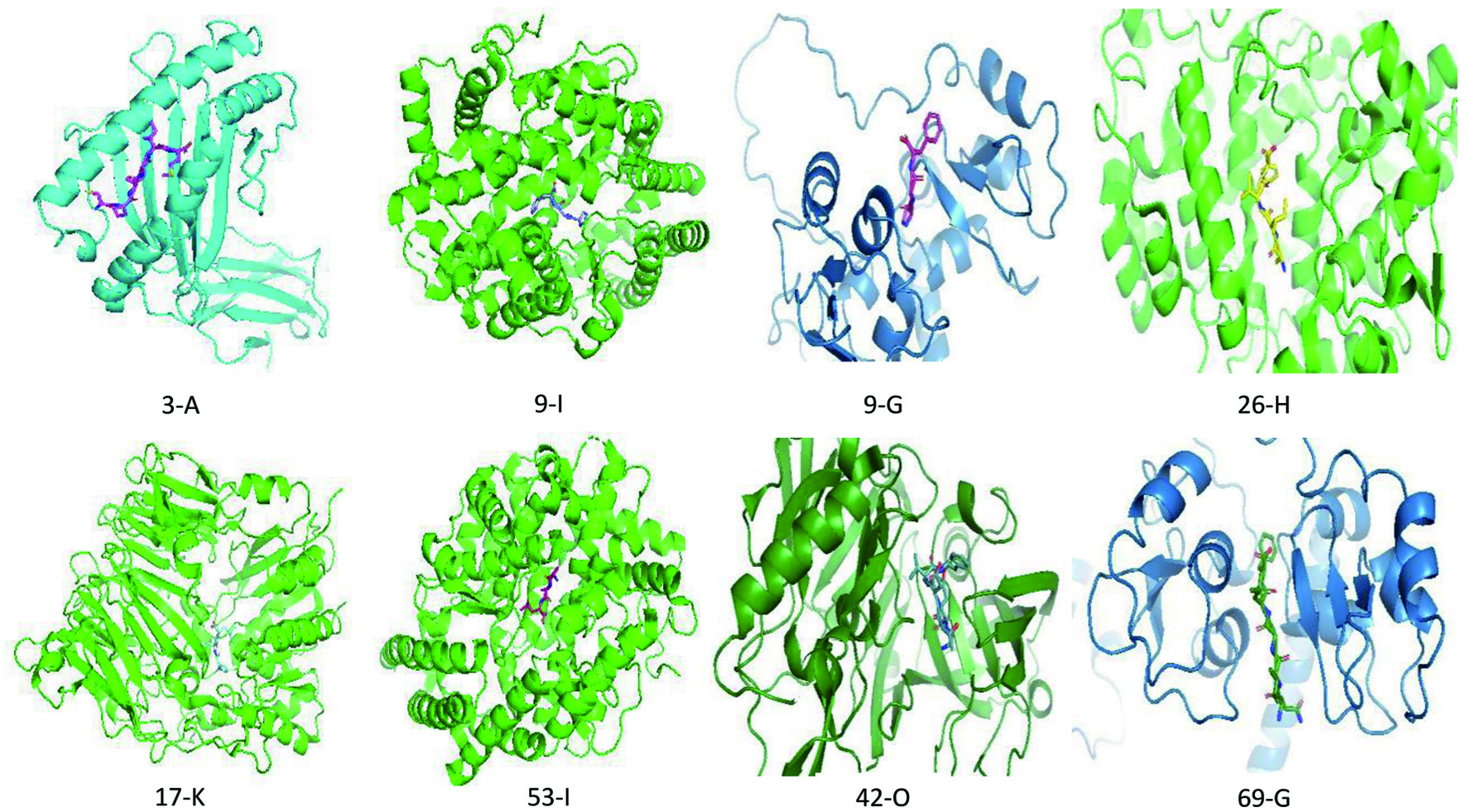
Tables
| Biological activity | |||||||
|---|---|---|---|---|---|---|---|
| Alpha-amylase inhibitor | ACE inhibitor | Antioxidative | Others | ||||
| Peptide | Sequence location | Peptide | Sequence location | Peptide | Sequence location | Peptide | Sequence location |
| *Anti-inflammatory, **Renin inhibitor, ***Anxiolytic-like peptide, ****Immuno-stimulating peptide, *****Stimulating vasoactive substance release. Bolded = reported in literature. Blue = multiple biological activities. Purple = component of other peptides of the same biological activity. | |||||||
| KLPGF | 64–68 | $pVSP$p | $p396–398$p | VHH | 21–23 | $bLPF$b* | $b239–241$b |
| $dEVSGL$d | $d 257–261$d | ILP | 116–118 | VHHANEN | 21–27 | $bYNL$b** | $b300–302$b |
| QITKPN | 92–97 | $pLVL$p | $p251–253$p | SALAM | 37–41 | $bYLG$b*** | $b43–45$b |
| $d AEAGVD$d | $d 356–361$d | LQP | 166–168 | $pLWE$p | $p190–192$p | GLF**** | 222–224 |
| $d EAGVD$d | $d 357–361$d | LLP | 253–255 | AEERYP | 110–115 | $dSSS$d ***** | $d318–320$d |
| NVLQPS | 164–169 | VFK | 15–17 | DEDTQAMP | 197–204 | ||
| LEPINF | 134–139 | LPG | 65–67 | $dGAA$d | $d 5–7$d | ||
| ANENIF | 24–29 | FCF | 11–13 | KG$pLWE$p | 188–192 | ||
| $bQIGLF$b | $b220–224$b | FFGR$dCVSP$d | 391–398 | LFC | 378–380 | ||
| HAEIN | 342–346 | ERKIKVYL | 284–291 | NEN | 25–27 | ||
| DHPFLF | 374–379 | FGR$dCVSP$d | 392–398 | $dSVL$d | $d304–306$d | ||
| NIFYCP | 27–32 | $bYNL$b | $b300–302b$ | ||||
| YAEERYPIL | 109–117 | $bYLG$b | $b43–45$b | ||||
| $bQIGLF$b | 220–224 | ||||||
| $bLPF$b | $b239–241$b | ||||||
| $dAVL$d | $d388–390$d | ||||||
| LFR | 223–225 | ||||||
| IFY | 28–30 | ||||||
| $dVVR$d | $d57–59$d | ||||||
| LKISQ | 332–336 | ||||||
| INKVVR | 54–59 | ||||||
| VYLPRM | 289–294 | ||||||
| KLP | 64–66 | ||||||
| INKV | 54–57 | ||||||
| $dSQAVH$d | $d335–339$d | ||||||
| QAVHA | 336–340 | ||||||
| LT$dSVL$dMA | 302–308 | ||||||
| SLR | 85–87 | ||||||
| $bYNL$b | $b300–302$b | ||||||
| $pLVL$pL | 251–254 | ||||||
| Biological activity | Peptide |
|---|---|
| Bolded = peptides with multiple biological activity. | |
| ACE inhibitor | $dVY$d, $dPR$d, $dAF$d, IF, $dGL$d, GR, $dSF$d, CF, $dEK$d, $dIL$d, $dAVL$d, $dVVR$d, $dVM$d, $dEF$d |
| Antioxidative | EL, $dVY$d |
| CaMPDE or Renin inhibitor | $dEF$d, $dSF$d |
| Glucose uptake stimulating peptide | $dVL$d, $dIL$d |
| Dipeptidyl peptidase IV inhibitor (DPP IV inhibitor) | $dEK$d, $dSL$d, $dGL$d, $dAF$d, $dIL$d, IN, $dPF$d, $dSF$d, SW, VH, $dVL$d, VN, $dVM$d, $dVY$d |
| Dipeptidyl peptidase III inhibitor (DPP-III inhibitor) | $dPR$d, $dPF$d, $dVY$d |
| Hypolipidemic peptide | $dEF$d |
| Regulator of phosphoglycerate kinase activity | $dSL$d |
| α | Hydrolysate peptides (Ligands) | % Probability of Predicted Targets | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | ||
| α indicates active peptide from serial number listed in the supplementary file Table s1, where peptides 1–65 were peptides obtained from enzymatic hydrolyzed protein, and peptide 66-71 were the most active peptides obtained from the unhydrolyzed protein screening. A: HLA class I histocompatibility antigen A-3 (P04439). B: Protein farnesyltransferase (P49354 P49356). C: Calpain 1 (P07384). D: Cathepsin (B and K or D) (P07858 or P07339). E: Complement factor B (P00751). F: Furin (P09958). G: Inhibitor of apoptosis protein 3 (E3 ubiquitin-protein ligase XIAP) (P98170). H: Cyclooxygenase-2 (P35354). I: Angiotensin-converting enzyme (P12821). J: Lipoxin A4 receptor (N-formyl peptide receptor 2) (by homology) (P25090). K: Dipeptidyl peptidase IV (P27487). L: Integrin alpha-V/beta-3 and alpha-5/beta-1 (P06756 P05106 and P05556 P08648). M: Neurokinin 1 receptor (by homology) (P25103). N: Mu/Delta opioid receptors (by homology) (P35372/P41143). O: Beta-secretase 1 (P56817). P: HMG-CoA reductase (P04035). Q: Tyrosyl-tRNA synthetase (P54577). R: Sodium/glucose cotransporter 1 (P13866). | |||||||||||||||||||
| 1 | GSIGAASM | 25 | |||||||||||||||||
| 3 | CPIAIM | 35 | 60 | ||||||||||||||||
| 4 | SAL | 25 | |||||||||||||||||
| 5 | GAK | 35 | 30 | 30 | |||||||||||||||
| 8 | VVR | 40 | 30 | ||||||||||||||||
| 9 | PGF | 40 | 35 | 25 | |||||||||||||||
| 11 | SSL | 25 | |||||||||||||||||
| 13 | QITK | 25 | |||||||||||||||||
| 17 | PIL | 75 | 50 | 75 | |||||||||||||||
| 20 | GGL | 70 | |||||||||||||||||
| 22 | QTAADQAR | 50 | |||||||||||||||||
| 24 | GIIR | 55 | |||||||||||||||||
| 26 | AIVF | 25 | 85 | 30 | 35 | 35 | 35/30 | ||||||||||||
| 29 | PVQM | 45 | |||||||||||||||||
| 30 | QIGL | 50 | 25 | 45 | |||||||||||||||
| 42 | GITDVF | 35 | 25 | ||||||||||||||||
| 44 | SGISSAESL | 60 | |||||||||||||||||
| 53 | AVL | 25 | 25 | 40 | 50 | 45 | 40 | ||||||||||||
| 55 | VY | 40 | 50 | 60 | |||||||||||||||
| 60 | IL | 25 | 25 | ||||||||||||||||
| 67 | YLG | 25 | 40/60 | ||||||||||||||||
| 68 | LPF | 90 | 30 | 50/30 | |||||||||||||||
| 69 | QIGLF | 55 | 25 | ||||||||||||||||
| 70 | VSP | 25 | 35 | 45 | |||||||||||||||
| 71 | LWE | 50 | |||||||||||||||||
| α | Hydrolysate peptides (Ligands) | Predicted Binding Energy ΔG (kcal.mol−1), Dissociation Constant Kd (10−7M) at 25.0 °C | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | |||
| α indicates active peptide from serial number listed in the supplementary file Table s1, where peptides 1–65 were peptides obtained from enzymatic hydrolyzed protein, and peptide 66–71 were the most active peptides obtained from the unhydrolyzed screening. A: HLA class I histocompatibility antigen A-3 (1AKJ). B: Protein farnesyltransferase (2H6I). C: Calpain 1 (AF-P07384-F1-model_v2). D: Cathepsin (B and K) (2PBH). E: Complement factor B (2OK5). F: Furin (6HLD). G: Inhibitor of apoptosis protein 3 (E3 ubiquitin-protein ligase XIAP) (AF-P98170-F1-model_v2). H: Cyclooxygenase-2 (5KIR). I: Angiotensin-converting enzyme (4C2N). J: Lipoxin A4 receptor (human formyl peptide receptor 2) (by homology) (6LW5). K: Dipeptidyl peptidase IV (3Q8W). L: Integrin alpha-V/beta-3 and alpha-5/beta-1 (4O02). M: Neurokinin 1 receptor (by homology) (6HLO). N: Mu/Delta opioid receptors (by homology) (AF-P35372-F1-model_v2). O: Beta-secretase 1 (2OHM). P: HMG-CoA reductase (AF-P04035-F1-model_v2). Q: Tyrosyl-tRNA synthetase (4QBT). R: Sodium/glucose cotransporter 1 (AF-P13866-F1-model_v2). AF represent Alphafold monomer v2.0 prediction obtained from UniProt structure section for each of the selected proteins, while structures of other proteins were obtained from RCSB protein databank. | ||||||||||||||||||||
| 1 | GSIGAASM | −8.5, 6.2 | ||||||||||||||||||
| 3 | CPIAIM | −8.3, 8.2 | −9.6, 0.93 | |||||||||||||||||
| 4 | SAL | −7.9, 17.0 | ||||||||||||||||||
| 5 | GAK | −7.5, 29.0 | −6.7, 120.0 | −7.7, 21.0 | ||||||||||||||||
| 8 | VVR | −7.2, 56.0 | −7.9, 17.0 | |||||||||||||||||
| 9 | PGF | −7.3, 42.0 | −6.5, 180.0 | −8.9, 2.9 | ||||||||||||||||
| 11 | SSL | −8.6, 5.0 | ||||||||||||||||||
| 13 | QITK | −5.9, 450.0 | ||||||||||||||||||
| 17 | PIL | −7.3, 42.0 | −7.7, 21.0 | −8.4, 7.2 | ||||||||||||||||
| 20 | GGL | −8.3, 8.2 | ||||||||||||||||||
| 22 | QTAADQAR | −8.9, 2.9 | ||||||||||||||||||
| 24 | GIIR | −8.9, 2.9 | ||||||||||||||||||
| 26 | AIVF | −8.8, 3.4 | −8.0, 13.0 | −8.1, 12.0 | −7.8, 19.0 | −5.6, 820.0 | −7.6, 27.0 | |||||||||||||
| 29 | PVQM | −8.3, 8.2 | ||||||||||||||||||
| 30 | QIGL | −7.5, 29.0 | −8.2, 9.3 | −8.5, 5.4 | ||||||||||||||||
| 42 | GITDVF | −8.4, 7.2 | −5.9, 460.0 | |||||||||||||||||
| 44 | SGISSAESL | −9.5, 1.1 | ||||||||||||||||||
| 53 | AVL | −6.8, 100.0 | −4.8, 2800.0 | −7.3, 45.0 | −8.0, 13.0 | −8.3, 8.2 | −8.9, 2.8 | |||||||||||||
| 55 | VY | −5.1, 1800.0 | −4.1, 9700.0 | −5.3, 1200.0 | ||||||||||||||||
| 60 | IL | −4.8, 2800.0 | −7.9, 17.0 | |||||||||||||||||
| 67 | YLG | −7.6, 27.0 | −5.7, 670.0 | |||||||||||||||||
| 68 | LPF | −7.2, 53.0 | −7.5, 29.0 | −6.2, 280.0 | ||||||||||||||||
| 69 | QIGLF | −8.0, 13.0 | −8.0, 13.0 | |||||||||||||||||
| 70 | VSP | −8.0, 13.0 | −5.2, 1500.0 | −6.7, 120.0 | ||||||||||||||||
| 71 | LWE | −8.0, 13.0 | ||||||||||||||||||
| α | Hydrolysate peptides (Ligands) | Predicted ADME Parameter | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MW | MR | TPSA (Å2) | Log P | ESOL Log S | ESOL Class | GIA | BBB | P-gp | CYPs Inhibitor | Log Kp (cm/s) | BS | SA | ||
| α indicates active peptide from serial number listed in the supplementary file Table s1, where peptides 1–65 were peptides obtained from enzymatic hydrolyzed protein, and peptide 66–71 were the most active peptides obtained from the unhydrolyzed screening. Legend: Physicochemical properties: Molecular weight (MW), Molar Refractivity (MR), Total polar surface area (TPSA). Lipophilicity: Consensus Log P. Water solubility: ESOL Log S, ESOL Class. Pharmacokinetics: Gastrointestinal absorption (GIA), Blood-brain barrier (BBB), P-glycoprotein (P-gp) substrate, Inhibition of Cytochrome P450 (CYPs) type CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4, Skin permeation (Log Kp). Drug-likeness: Bioavailability Score (BS), Medicinal chemistry: Synthetic accessibility (SA). | ||||||||||||||
| 1 | GSIGAASM | 692.78 | 167.31 | 332.78 | −3.05 | 1.38 | Highly soluble | Low | No | Yes | No | −14.58 | 0.17 | 6.10 |
| 3 | CPIAIM | 646.86 | 173.61 | 264.13 | 0.4 | −0.78 | Very soluble | Low | No | No | No | −11.99 | 0.17 | 6.06 |
| 4 | SAL | 289.33 | 71.44 | 141.75 | −1.39 | 1.14 | Highly soluble | Low | No | No | No | −10.44 | 0.55 | 3.20 |
| 5 | GAK | 274.32 | 68.18 | 147.54 | −1.54 | 1.79 | Highly soluble | Low | No | No | No | −10.91 | 0.55 | 2.89 |
| 8 | VVR | 372.46 | 98.61 | 183.42 | −1.14 | 1.57 | Highly soluble | Low | No | Yes | No | −11.72 | 0.55 | 3.92 |
| 9 | PGF | 319.36 | 87.05 | 107.53 | −0.22 | 0.29 | Highly soluble | High | No | No | No | −10.18 | 0.55 | 2.85 |
| 11 | SSL | 305.33 | 72.60 | 161.98 | −2 | 1.97 | Highly soluble | Low | No | No | No | −11.51 | 0.55 | 3.24 |
| 13 | QITK | 488.58 | 123.32 | 239.96 | −1.65 | 1.67 | Highly soluble | Low | No | Yes | No | −12.91 | 0.17 | 4.90 |
| 17 | PIL | 341.45 | 96.21 | 107.53 | 0.59 | −0.2 | Very soluble | High | No | Yes | No | −9.55 | 0.55 | 3.69 |
| 20 | GGL | 245.28 | 60.67 | 121.52 | −1.1 | 1.32 | Highly soluble | High | No | No | No | −10.15 | 0.55 | 2.31 |
| 22 | QTAADQAR | 859.88 | 204.08 | 472.63 | −5.64 | 3.13 | Highly soluble | Low | No | Yes | No | −18.22 | 0.17 | 7.25 |
| 24 | GIIR | 457.57 | 120.84 | 212.52 | −0.51 | 0.05 | Highly soluble | Low | No | Yes | No | −10.75 | 0.17 | 4.69 |
| 26 | AIVF | 448.56 | 121.8 | 150.62 | 1.2 | −1.4 | Very soluble | Low | No | Yes | No | −9.46 | 0.55 | 4.22 |
| 29 | PVQM | 473.59 | 124.13 | 205.02 | −1.3 | 1.23 | Highly soluble | Low | No | No | No | −12.44 | 0.17 | 4.66 |
| 30 | QIGL | 429.51 | 109.84 | 193.71 | −0.67 | 1.07 | Highly soluble | Low | No | Yes | No | −11.68 | 0.17 | 4.14 |
| 42 | GITDVF | 650.72 | 164.39 | 266.35 | −0.62 | −1.07 | Very soluble | Low | No | Yes | No | −11.75 | 0.11 | 5.69 |
| 44 | SGISSAESL | 849.88 | 200.46 | 414.34 | −4.72 | 1.99 | Highly soluble | Low | No | Yes | No | −16.81 | 0.11 | 7.02 |
| 53 | AVL | 301.38 | 79.89 | 121.52 | −0.08 | 0.72 | Highly soluble | High | No | No | No | −10.13 | 0.55 | 3.25 |
| 55 | VY | 280.32 | 74.56 | 112.65 | 0.01 | 0.25 | Highly soluble | High | No | No | No | −9.80 | 0.55 | 2.64 |
| 60 | IL | 244.33 | 67.28 | 92.42 | 0.49 | 0.25 | Highly soluble | High | No | No | No | −9.00 | 0.55 | 2.87 |
| 67 | YLG | 351.40 | 91.98 | 141.75 | −0.02 | 0.14 | Highly soluble | Low | No | No | No | −10.26 | 0.55 | 3.04 |
| 68 | LPF | 375.46 | 106.28 | 112.73 | 0.85 | −0.86 | Very soluble | High | No | Yes | No | −9.51 | 0.55 | 3.52 |
| 69 | QIGLF | 576.68 | 151.75 | 222.81 | 0.16 | −0.55 | Very soluble | Low | No | Yes | No | −11.53 | 0.17 | 5.00 |
| 70 | VSP | 301.34 | 78.14 | 132.96 | −1.41 | 1.53 | Highly soluble | Low | No | Yes | No | −11.20 | 0.55 | 3.27 |
| 71 | LWE | 446.5 | 118.01 | 174.61 | 0.41 | −0.95 | Very soluble | Low | No | No | No | −10.09 | 0.11 | 4.06 |
| α | Hydrolysate peptides (Ligands) | Predicted Physicochemical Parameter | ||||||
|---|---|---|---|---|---|---|---|---|
| Toxicity | Hemolytic (SVM score) | pI | Hydrophobicity | Hydropathicity | Hydrophilicity | Charge | ||
| α indicates active peptide from serial number listed in the supplementary file Table s1, where peptides 1–65 were peptides obtained from enzymatic hydrolyzed protein, and peptide 66–71 were the most active peptides obtained from the unhydrolyzed screening. ToxinPred webserver based on support vector machine (SVM) score: positive (YES) or negative (NO). Hemolytic prediction based on SVM score. | ||||||||
| 1 | GSIGAASM | No | 0.49 | 5.88 | 0.16 | 0.95 | −0.44 | 0.00 |
| 3 | CPIAIM | No | 0.49 | 5.85 | 0.32 | 2.27 | −1.07 | 0.00 |
| 4 | SAL | No | 0.49 | 5.88 | 0.17 | 1.60 | −0.67 | 0.00 |
| 5 | GAK | No | 0.49 | 9.11 | −0.23 | −0.83 | 0.83 | 1.00 |
| 8 | VVR | No | 0.49 | 10.11 | −0.23 | 1.30 | 0.00 | 1.00 |
| 9 | PGF | No | 0.49 | 5.88 | 0.23 | 0.27 | −0.83 | 0.00 |
| 11 | SSL | No | 0.49 | 5.88 | 0.00 | 0.73 | −0.40 | 0.00 |
| 13 | QITK | No | 0.49 | 9.11 | −0.31 | −0.90 | 0.25 | 1.00 |
| 17 | PIL | No | 0.49 | 5.88 | 0.40 | 2.23 | −1.20 | 0.00 |
| 20 | GGL | No | 0.49 | 5.88 | 0.28 | 1.00 | −0.60 | 0.00 |
| 22 | QTAADQAR | No | 0.49 | 6.19 | −0.41 | −1.29 | 0.56 | 0.00 |
| 24 | GIIR | No | 0.49 | 10.11 | −0.03 | 1.02 | −0.15 | 1.00 |
| 26 | AIVF | No | 0.49 | 5.88 | 0.53 | 3.33 | −1.57 | 0.00 |
| 29 | PVQM | No | 0.49 | 5.88 | 0.01 | 0.25 | −0.65 | 0.00 |
| 30 | QIGL | No | 0.49 | 5.88 | 0.18 | 1.10 | −0.85 | 0.00 |
| 42 | GITDVF | No | 0.49 | 3.80 | 0.19 | 1.15 | −0.53 | −1.00 |
| 44 | SGISSAESL | No | 0.49 | 4.00 | 0.00 | 0.33 | 0.01 | −1.00 |
| 53 | AVL | No | 0.49 | 5.88 | 0.44 | 3.27 | −1.27 | 0.00 |
| 55 | VY | No | 0.49 | 5.88 | 0.28 | 1.45 | −1.90 | 0.00 |
| 60 | IL | No | 0.49 | 5.88 | 0.63 | 4.15 | −1.80 | 0.00 |
| 67 | YLG | No | 0.49 | 5.88 | 0.24 | 0.70 | −1.37 | 0.00 |
| 68 | LPF | No | 0.49 | 5.88 | 0.36 | 1.67 | −1.43 | 0.00 |
| 69 | QIGLF | No | 0.49 | 5.88 | 0.27 | 1.44 | −1.18 | 0.00 |
| 70 | VSP | No | 0.49 | 5.88 | 0.07 | 0.60 | −0.40 | 0.00 |
| 71 | LWE | No | 0.49 | 4.00 | 0.09 | −0.20 | −0.73 | −1.00 |
| α | Hydrolysate peptides (Ligands) | Antimicrobial* | Antioxidant** | Anticancer*** | Anti-inflammatory**** | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibacterial | Antiviral | Antifungal | |||||||||
| α indicates active peptide from serial number listed in the supplementary file Table s1, where peptides 1–65 were peptides obtained from enzymatic hydrolyzed protein, and peptide 66–71 were the most active peptides obtained from the unhydrolyzed screening. * = 0.5 Probability cut-off. ** = 0.5 Probability cut-off. *** = SVM score: positive (YES) or negative (NO). | |||||||||||
| 1 | GSIGAASM | YES | NO | YES | NO | NO | LOW | ||||
| 3 | CPIAIM | YES | YES | YES | NO | YES | LOW | ||||
| 4 | SAL | NO | NO | NO | NO | YES | NO | ||||
| 5 | GAK | YES | YES | YES | NO | NO | NO | ||||
| 8 | VVR | YES | YES | NO | NO | YES | NO | ||||
| 9 | PGF | YES | YES | YES | NO | YES | NO | ||||
| 11 | SSL | YES | NO | NO | NO | YES | NO | ||||
| 13 | QITK | YES | YES | NO | NO | YES | NO | ||||
| 17 | PIL | YES | YES | YES | NO | YES | NO | ||||
| 20 | GGL | YES | YES | YES | NO | NO | NO | ||||
| 22 | QTAADQAR | NO | NO | NO | NO | YES | LOW | ||||
| 24 | GIIR | YES | YES | YES | NO | NO | MEDIUM | ||||
| 26 | AIVF | YES | NO | NO | NO | YES | LOW | ||||
| 29 | PVQM | YES | YES | NO | NO | YES | NO | ||||
| 30 | QIGL | YES | YES | YES | NO | YES | LOW | ||||
| 42 | GITDVF | NO | NO | NO | NO | NO | NO | ||||
| 44 | SGISSAESL | NO | NO | NO | NO | YES | LOW | ||||
| 53 | AVL | YES | YES | NO | NO | YES | NO | ||||
| 55 | VY | YES | YES | NO | NO | YES | NO | ||||
| 60 | IL | YES | YES | YES | NO | YES | NO | ||||
| 67 | YLG | YES | NO | NO | NO | YES | NO | ||||
| 68 | LPF | YES | YES | YES | NO | YES | NO | ||||
| 69 | QIGLF | YES | NO | YES | NO | YES | MEDIUM | ||||
| 70 | VSP | NO | NO | NO | NO | YES | NO | ||||
| 71 | LWE | YES | YES | NO | NO | YES | NO | ||||
| **** Confidence | Score range | Sensitivity | Specificity | ||||||||
| α indicates active peptide from serial number listed in the supplementary file Table s1, where peptides 1–65 were peptides obtained from enzymatic hydrolyzed protein, and peptide 66–71 were the most active peptides obtained from the unhydrolyzed screening. * = 0.5 Probability cut-off. ** = 0.5 Probability cut-off. *** = SVM score: positive (YES) or negative (NO). | |||||||||||
| High | Score ≥ 0.468 | 63.22% | 90.30% | ||||||||
| Medium | 0.468 > Score ≥ 0.388 | 71.90% | 80.11% | ||||||||
| Low | 0.388 >Score ≤ 0.342 | 78.39% | 70.87% | ||||||||