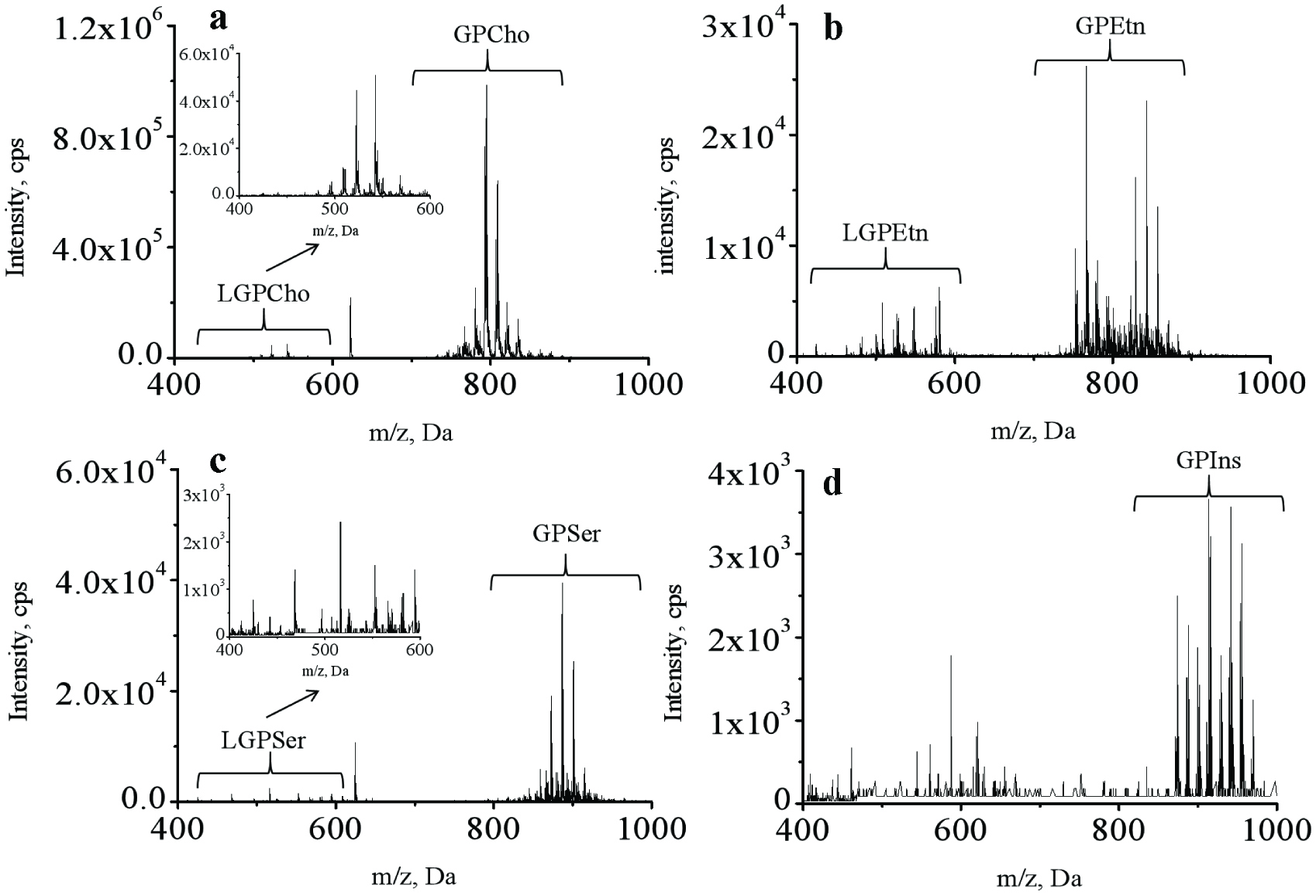
Figure 1.
Specific detection of glycerophospholipids in lipids recovered from different body parts of sea cucumber by using direct infusion mass spectrometric approaches.
a–d, first-stage MS spectra of glycerophosphocholines/lysoglycerophosphocholines (GPCho/LGPCho), glycerophosphoethanolamines/lysoglycerophosphoethanolamines (GPEtn/LGPEtn), glycerophosphoserines/lysoglycerophosphoserines (GPSer/LGPSer) and glycerophosphatidylinositols (GPIns), respectively.