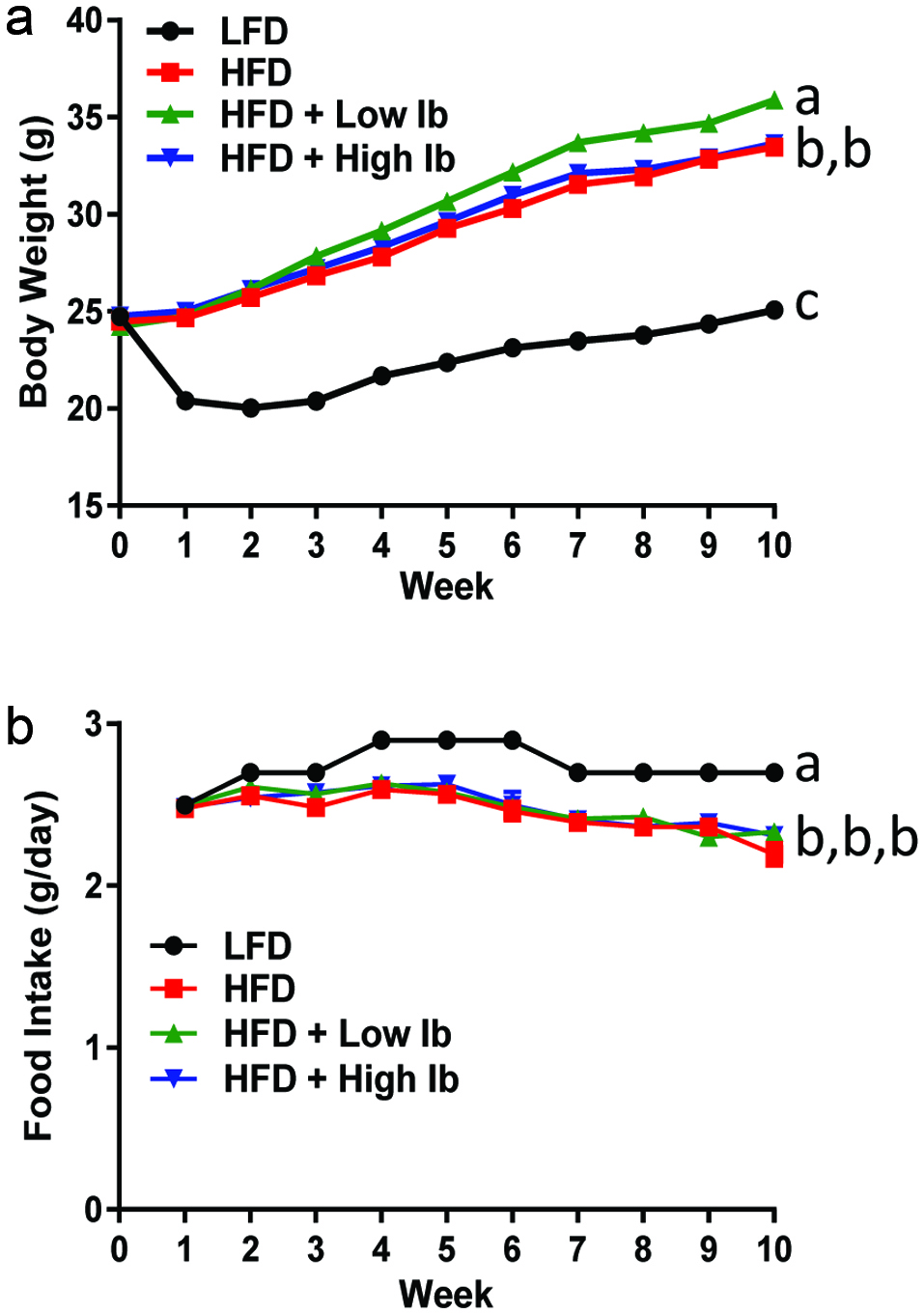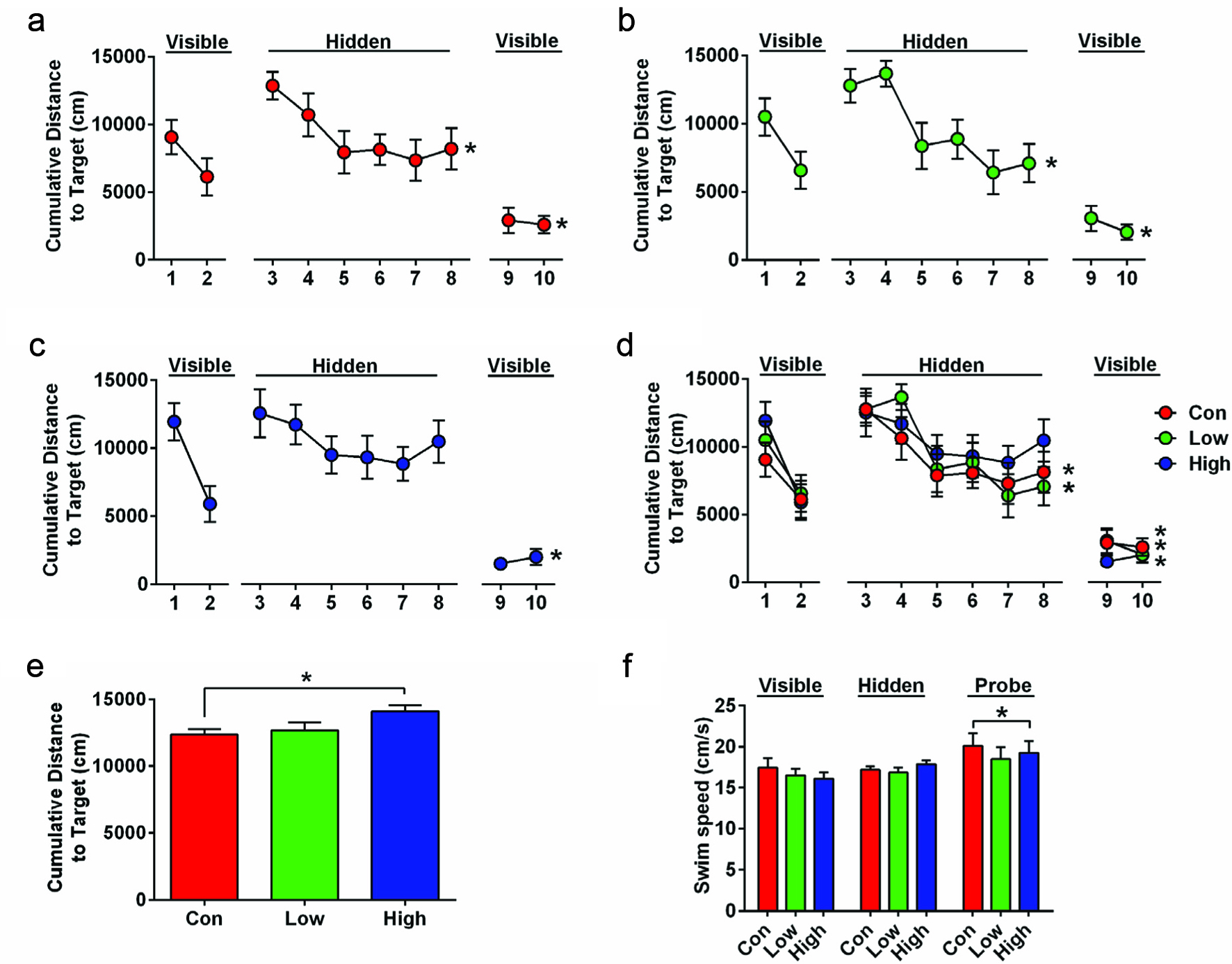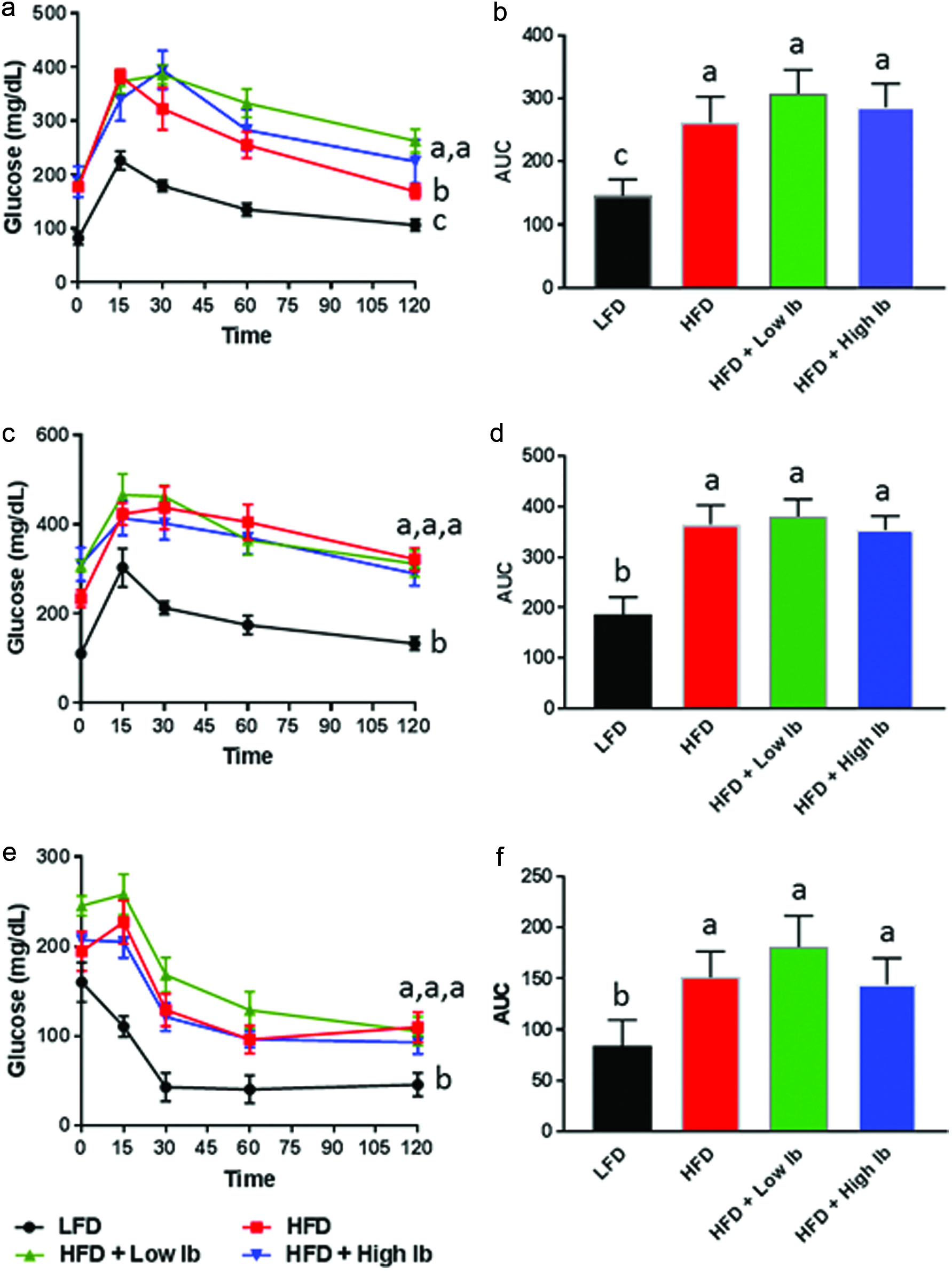
Influence of diet and iboga extract on glucose tolerance in mice at 4 weeks (a, b) and 9 weeks (c, d) and on insulin tolerance at 9 weeks (e, f) of the feeding study.
The mice were fasted for 6 h before i.p. injection of glucose, 2 g/kg body weight, or of insulin, 0.75 U/kg body weight, before blood glucose was analyzed at designated times. LFD, low-fat diet; HFD, high-fat diet; Ib, ibogaine. Data presented in panels A, C, and E were analyzed using a repeated-measures-in-time design ANOVA in PROC MIXED for repeated measures within animals (n = 5 per group). Data presented in panels B, D, and F were analyzed using a one-way ANOVA and a post-hoc Tukey’s multiple comparison test. Statistical differences between groups are indicated with different letters (a, b, c; p < 0.05).