| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Review
Volume 18, June 2022, pages 43-51
Fate of dietary phytochemicals in human gut and interaction with intestinal flora
Bin Dua, #, Zilong Maa, #, Guang Xinb, Yuedong Yanga, *, Baojun Xuc, *
aHebei Key Laboratory of Natural Products Activity Components and Function, Hebei Normal University of Science and Technology, Qinhuangdao, Hebei 066004, China
bCollege of Food Science, Shenyang Agricultural University, Shenyang, Liaoning 110866, China
cFood Science and Technology Program, BNU–HKBU United International College, Zhuhai, Guangdong 519087, China
*Corresponding author: Yuedong Yang, Hebei Key Laboratory of Natural Products Activity Components and Function, Hebei Normal University of Science and Technology, Qinhuangdao, Hebei 066004, China. E-mail: kycyyd@126.com; Baojun Xu, Food Science and Technology Program, BNU-HKBU United International College, Zhuhai, Guangdong 519087, China. E-mail: baojunxu@uic.edu.cn
DOI: 10.31665/JFB.2022.18307
Received: March 26, 2022
Revised received & accepted: June 23, 2022
| Abstract | ▴Top |
With the deterioration of the environment and the improvement of human self-protection awareness, the voice of human beings for returning to nature is increasing. There is an increasing interest in regulating the composition and metabolic function of the gastrointestinal microbial community with natural plant diet methods to improve health and prevent or treat diseases. As the “invisible organ” of human beings, the intestinal flora is closely related to human health. Dysbacteriosis can induce a variety of diseases. The regulatory effect of natural plant ingredients on the intestinal flora is expected to be a new treatment for related diseases. The metabolic mechanism of several main components of natural plants in the human body and their influence on the intestinal flora were presented in this paper. Meanwhile, the mechanism of action of intestinal flora in several typical diseases was introduced. In addition, this paper gathered and analyzed previous studies in order to gain insight how plant components can improve diseases by regulating intestinal flora.
Keywords: Phytochemicals; Gut microbiota; Oxidative stress; Reactive oxygen species; Obesity; Neurological diseases; Intestinal diseases
| 1. Introduction | ▴Top |
A number of mixtures of pure bioactive compounds or phytochemical-containing plant extracts provide synergy with regard to antioxidant status, anti-inflammation, anti-cancer and chemoprevention of several oxidative stress and metabolic disorders in vitro (Phan et al., 2018). Intestinal flora is the general term for a variety of microorganisms that are parasitic in the human intestine (Shepherd et al., 2018). The population of microorganisms that settle in the human intestine is very complex. The number of cells is 10 times that of our own cells. Each gram of intestinal contents contains about 1012 microbial cells and 1,000 different species (Knight and Girling, 2003), weighing about 2 kg (De Vos and de Vos, 2012; De Vos and Nieuwdorp, 2013). The intestinal flora plays an important role in the normal function of the host organism. Different intestinal bacteria can perform different functions. For example, glycolytic bacteria such as Bacteroides and Bifidobacteria can metabolize carbohydrates, Streptococcus, Staphylococcus, etc. Proteolytic bacteria can obtain energy from protein fermentation, and the metabolic activity of bacteria is mainly defined by its genome and epigenome (Kashtanova et al., 2016). The collective genome of these gut bacteria is called the “microbiome”, and the number of genes is at least one hundred times that of the human genome (Qin et al., 2010). More than 90% of the human colon is Firmicutes and Bacteroidetes, followed by Actinomyces, Proteus, Fusion Bacillus and Verrucous microorganisms (Robles Alonso and Guarner, 2013). The relationship between these microorganisms and our body is diverse, some are good for health, and the other is harmful. The composition of the flora depends on many factors, such as genetic factors and eating habits. With the introduction of high-throughput sequencing technology, we have a further understanding of the relationship between the bacterial community of the microbiota and human health (Kowalska and Olejnik, 2016).
The intestinal flora is closely related to human health and plays an important role in the metabolism and immune system, as shown in Figure 1. It is called the “invisible organ of the human body”. Disorders of the intestinal flora are related to many diseases, such as: inflammatory bowel disease (IBD), irritable bowel syndrome, autism, obesity, type-2 diabetes, etc. Studies have shown that the ratio of Firmicutes and Bacteroides in obese people is low, and the abundance of Firmicutes and Clostridia in patients with type-2 diabetes is low (Ma et al., 2019), and Escherichia and Streptococcus are abundant in people with cardiovascular diseases. The relative abundance of Lactobacillus, Faebacterium, Bifidobacterium, and Collins is relatively low in patients with gastrointestinal diseases (Song et al., 2019), and in patients with neurological diseases, Bifidobacterium is lower and Firmicutes is higher. A large number of studies have revealed the relationship between intestinal flora and these diseases. This paper reviews several typical diseases. The correlation between Alzheimer’s disease and intestinal flora reveals the impact of intestinal flora on the nervous system, which is one of the hotspots of current research. Obesity, as one of the world’s major health problems, can also cause other chronic diseases. Intestinal flora is closely related to obesity. Natural and harmless weight-loss products are the universal needs of obese patients today. Inflammatory bowel disease often recurs illness protraction. Affected by many factors such as eating habits and intestinal flora, there is no effective cure so far. The regulatory effect of plant ingredients on the intestinal flora is expected to alleviate the pain of IBD patients. Cardiovascular disease, also known as circulatory system disease, is a series of diseases involving the circulatory system, including high blood pressure, hyperlipidemia, coronary heart disease and many other diseases. There are often no obvious symptoms in the early stage, but there is a fatal risk. The intake of active ingredients in plants can effectively reduce the risk of cardiovascular disease (Latif et al., 2021).
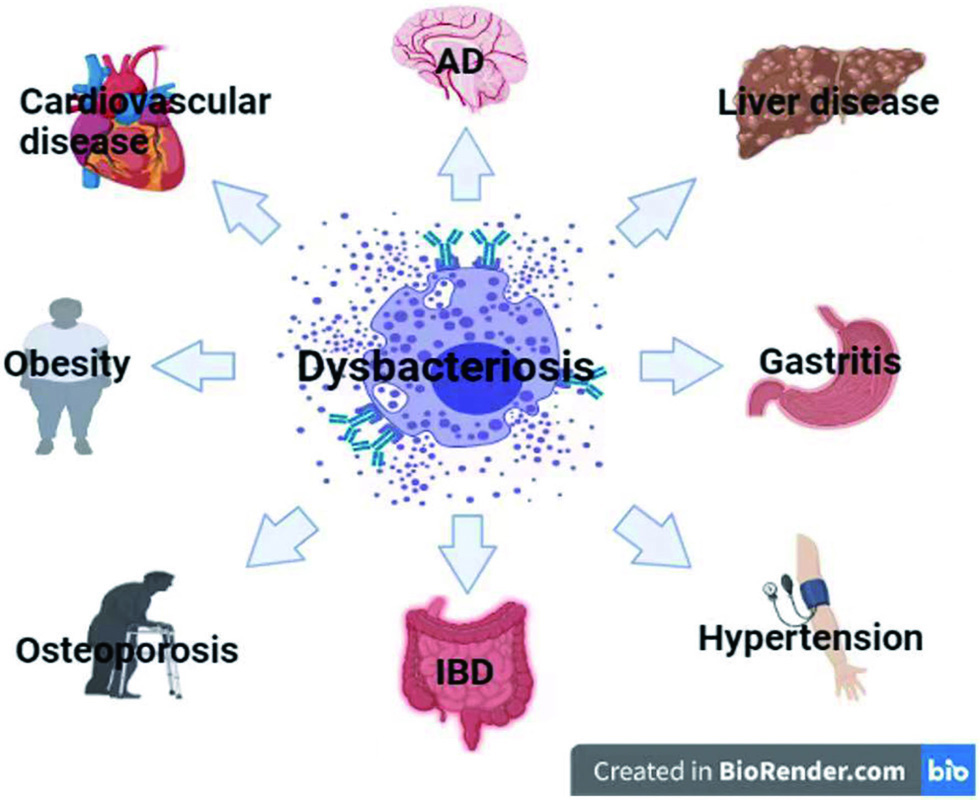 Click for large image | Figure 1. Dysbacteriosis and related diseases. This figure was created with icons provided by bio render (https://biorender.com). |
Plant ingredients are an important part of natural medicines and play an important role in the fight against diseases. Plant components have novel structure, many types, high curative effect, less adverse reactions and high screening hit rate. Many drugs currently used are directly or indirectly derived from natural products (mainly plant ingredients). Typical examples are berberine (Ortiz et al., 2014), an anti-gastrointestinal inflammatory drug, and artemisinin (Talman et al., 2019), an antimalarial drug. It has always been a goal pursued by medical workers to find effective ingredients from medicinal plants, especially botanicals with a basis for clinical application, and optimize the structure through structural modification, synthesis, and biotransformation, and then to develop new drugs. The research of phytochemical constituents has greatly promoted the development of pharmaceutical chemistry and become an important source for the discovery of new drugs or drug active lead compounds.
The role of plant active ingredients in the human body and the way of transportation are very complicated. In the process of food digestion, many food chemical components (especially plant foods) are not digested and absorbed in the small intestine, but reach the colon and are metabolized by intestinal microbes (Rinninella et al., 2019). The main chemical components of plant foods are water, carbohydrates, proteins, lipids, polypeptides, amino acids, pectins, cellulose, hemicellulose, starch and some free metal ions, and these components interact with the intestinal flora. This paper selects three main active ingredients (polysaccharides, proteins, phenols) that are relatively abundant in plants to review, and introduces the transport pathways of these substances in the body and their effects on the intestinal flora. At the same time, the relationship between the intestinal flora and related diseases is reviewed, which provides new ideas for the treatment of these diseases and the rational use of plant resources.
| 2. The influence of dietary components on the intestinal flora | ▴Top |
2.1. Carbohydrates (polysaccharides)
The indigestible complex polysaccharides in the diet are the main carbon source of the intestinal flora (Bäckhed et al., 2005). After dietary polysaccharides reach the colon, the intestinal flora will degrade the polysaccharides into monosaccharides or oligosaccharides through different degradation systems and transport systems, and then transport these monosaccharides or oligosaccharides into the cell for further degradation and fermentation (Dalile et al., 2019). Therefore, these polysaccharides are called fermented polysaccharides, and there are three main categories: non-starch polysaccharides, resistant starch and dietary fiber. Fermented polysaccharides can produce gas, heat and short-chain fatty acids under the joint action of intestinal flora. In addition to polysaccharides in food, polysaccharides shed by intestinal epithelial cells are also a stable source of nutrients for the microbiota (Koropatkin et al., 2012). In the intestinal flora, Bacteroides is the dominant strain for degrading polysaccharides, and 20% of its genome is used to complete the degradation of polysaccharides (Singh, 2019). Humans discovered the polysaccharide utilization site (PUL) for the first time in Bacteroides intestinal tract (Reichardt et al., 2018). PUL encodes different glycoside hydrolases (Luis et al., 2018), which have been determined to decompose different substrates. In addition to cellulose, all polysaccharides entering the intestine have corresponding polysaccharide transport systems to be responsible for the transportation and decomposition of polysaccharides. These systems and related functional regulatory proteins are collectively called starch utilization systems (Tancula et al., 1992). Taking Bacteroides as an example, as shown in Figure 2, polysaccharides are initially combined with surface glycan binding proteins (SGBPs), and the original polysaccharides are decomposed into multiple oligosaccharides by the outer membrane surface glycoside hydrolase (GH) and then bound by related glycan binding proteins (SusD) Binding, transported from the outer membrane into the periplasmic space by the outer membrane transporter, these oligosaccharides are further broken down into smaller oligosaccharides through the GH or polysaccharide hydrolase (PL) in the periplasmic space (Foley et al., 2016), and then pass through the inner membrane. The membrane transporter transports the enzymatic hydrolysis products into the cell. In addition to Gram-negative bacteria, the Firmicutes phylum has an outstanding ability to utilize polysaccharides. Although Firmicutes encodes fewer carbohydrate degrading enzymes, it encodes more ABC transporters (ATP binding transporters). To transport carbohydrates (Mahowald et al., 2009). In addition, the Bifidobacteria in Actinomycetes can also degrade part of polysaccharides (Koropatkin et al., 2012).
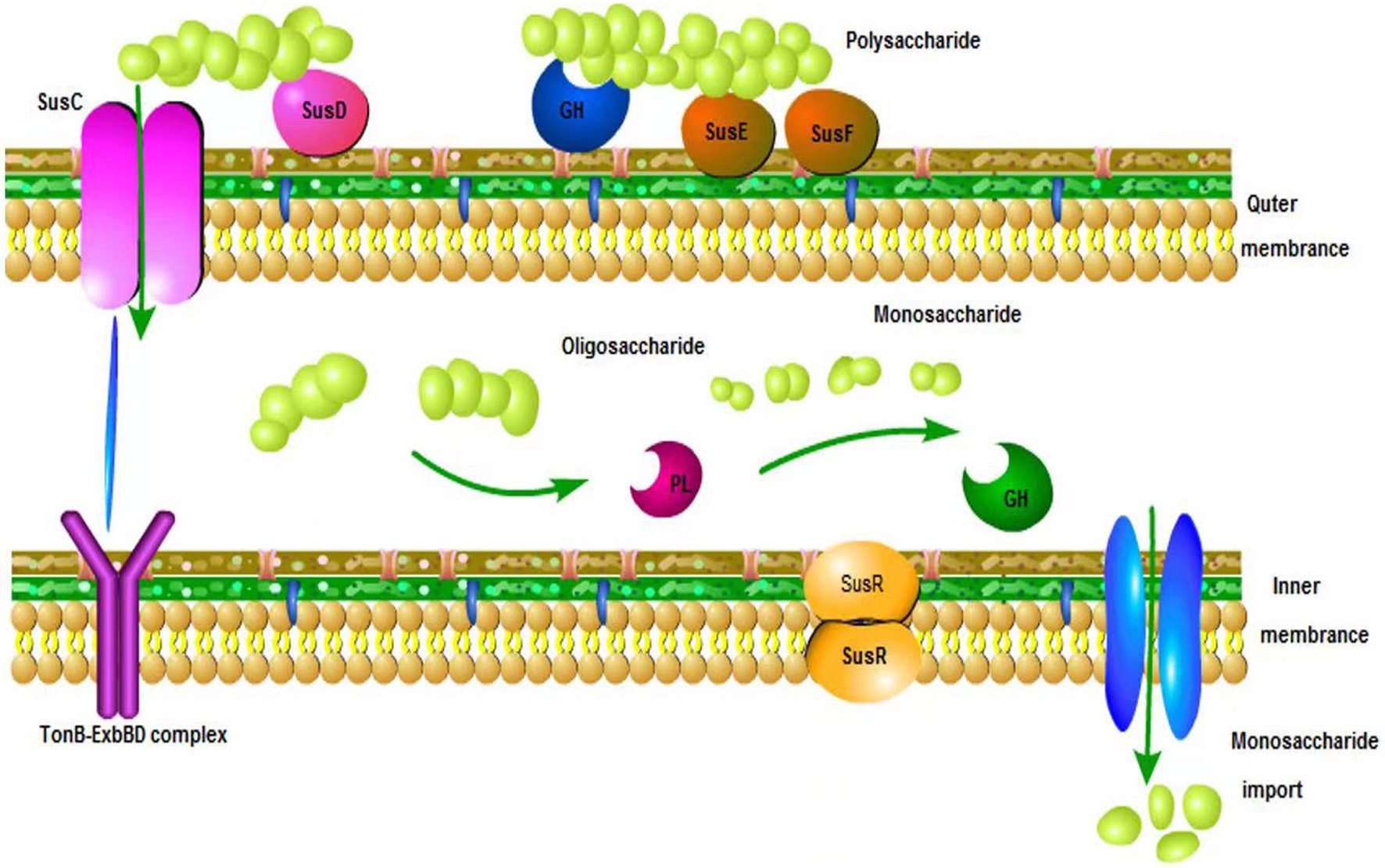 Click for large image | Figure 2. Model of polysaccharide utilization system in the Bacteroides. |
In general, the more complex the polysaccharide, the more glycosidase are necessary to metabolize it (Martens et al., 2011). Some bacteria possess many different enzymes that allow them to metabolize dozens of different complex carbohydrates, while other microbes are only able to utilize one or a few different polysaccharides. For example, Bacteroides thetaiotaomicron and B. ovatus, bacteria found in the human microbiota, are capable of metabolizing more than a dozen different types of glycans (Scott et al., 2014). Different short chain fatty acids produced after polysaccharide degradation also have important effects on human body. The main bacterial fermentative end products of complex carbohydrates are short chain fatty acids (SCFAs), namely acetate, propionate, and butyrate. Butyrate is the key energy source for colonocytes and enterocytes. Propionate can be utilized locally through conversion into glucose by intestinal gluconeogenesis or diffuse into the portal vein to be utilized as a substrate for hepatic gluconeogenesis (Cummings et al., 1987). Between 90 and 99% of SCFAs are absorbed in the gut or used by the microbiota (Ruppin et al., 1980). A large number of studies have proved that SCFAs influence gastrointestinal epithelial cell integrity, glucose homeostasis, lipid metabolism, appetite regulation, and immune function (Perry et al., 2016). It can also down regulate proinflammatory cytokines (Chang et al., 2014), affect gene expression and cell differentiation (Fung et al., 2012).
In addition, increased propionate flux through the liver has been shown to reduce intrahepatic triglyceride which is also likely to elicit an improvement in hepatic and whole-body glucose homeostasis (Chambers et al., 2015). Furthermore, acetate has been linked to suppression of adipocyte lipolysis, thus reducing free fatty acid (FFA) flux to the liver mitigating against fatty liver induced deterioration in glucose homeostasis (Crouse et al., 1968). SCFA also inhibit insulin stimulated lipid accumulation in adipocytes via FFAR 2 signaling, resulting in small more responsive adipocytes which is associated with reduced adipose inflammatory infiltrate (Kimura et al., 2013). Acetate also appears to stimulate leptin secretion in adipocytes (Zaibi et al., 2010). Leptin is an important adipose derived homeostatic signal which regulates energy balance and appetite (Halaas et al., 1995).
2.2. Peptides (proteins)
Protein in food is also the main nitrogen source of intestinal microbes. Exfoliated small intestine and large intestine mucosal cells and digestive enzyme proteins secreted into the lumen of the small intestine are also stable nitrogen sources of intestinal microbes. The intestinal flora has an efficient protein metabolism mechanism(Hamer et al., 2012). By participating in the body’s protein metabolism, the intestinal flora provides a nitrogen source for its own growth on the one hand, and also helps the host to decompose and synthesize essential amino acids to meet physiological needs (Morowitz et al., 2011). Food and endogenous proteins are hydrolyzed into peptides and amino acids by proteases and peptidases produced by the host and bacteria, and the released amino acids are used by the host and microorganisms. Amino acids can be metabolized into many microbial metabolites, which are involved in various physiological functions related to host health and disease. The intestinal flora mainly uses amino acids in the lumen of the small intestine. The most abundant amino acid-fermenting bacteria in the small intestine is the Clostridium spp (Dai et al., 2011). When the bacteria take in amino acids, the amino acids can directly become bacterial cell components, or continue to be catabolized. Some amino acids enter the large intestine and are used by bacteria (Davila et al., 2013). The regulatory effect of protein on the intestinal flora is two-way. For example, the components of breast milk protein are conducive to the survival and reproduction of Bifidobacteria, while milk and legume proteins are the opposite. The lysozyme contained in breast milk can affect the cell wall of bacteria(Wu et al., 2020), leading to the depolymerization of bacterial cell membrane mucopolysaccharides. Bifidobacteria and Lactobacilli carrying lysozyme can enhance the continuous digestion and absorption of the products after the lysis of these bacteria through trypsin. The high concentration of calcium and casein in milk are fatty acid salts that have the properties of inhibiting Bifidobacterium, and therefore have a negative impact on Bifidobacteria.
The primary bacteria related to protein metabolism in the small intestine include Klebsiella, Escherichia coli, Streptococcus, Vibrio dextrinus, Vibrio photoalgae and Vibrio fatty liver anaerobes. Some of these bacteria can directly metabolize amino acids and secrete various proteases and polypeptide enzymes (Fan et al., 2017). Dietary proteins are digested into amino acids and oligopeptides by proteases and many peptidases, and then absorbed in the epithelium of the small intestine (Bos et al., 2005). In addition, highly digestible proteins may partially escape the digestion of the small intestine, and a large amount of nitrogen-containing substances may be transferred from the small intestine to the large intestine (Evenepoel et al., 1999). Undigested proteins and peptides are hydrolyzed by intestinal microorganisms and residual pancreatic proteases to produce a large number of microbial metabolites (Blachier et al., 2007). These are mainly SCFAs, ammonia, polyamines and hydrogen sulfide. Some of them can be transported to colon cells and have beneficial or harmful effects on these epithelial cells according to their concentration in the lumen. Some bacterial metabolites are transported to portal vein blood, which can produce various physiological effects on liver and surrounding organs and tissues (Davila et al., 2013).
The source, concentration and amino acid balance of dietary protein are the main factors affecting the composition, structure and function of intestinal microorganisms. Proteins mainly come from plants or animals. Compared with animal protein, the price of plant protein is usually lower and has some advantages in food safety. Both soybean and peanut proteins have been shown to play a positive role in regulating beneficial intestinal bacterial components (Peng et al., 2015). For example, peanut protein can increase the abundance of Bifidobacteria and reduce intestinal pH, thereby inhibiting toxic metabolite products. However, protein intake is also closely related to health. Too much protein in the human diet increases the risk of colon disease. Residual nitrogenous compounds not absorbed by the small intestine will be transferred to the distal intestine and metabolized by microorganisms in this part of the gastrointestinal tract. The quantity and types of microbial metabolites are affected by dietary protein intake. Some microbial metabolites are toxic, such as hydrogen sulfide, ammonia and indole compounds, which may have a negative impact on host health. High concentration of protein supplementation will destroy the steady state of ecosystem, reduce the number of beneficial microorganisms and increase the number of potential pathogens (Zhao et al., 2019). When the protein concentration is too low to meet the basic needs of the host, it will increase the abundance of potential pathogens and reduce the number of prebiotics. Amino acid balance is also the main factor affecting human protein digestibility. The imbalance of amino acid ratio in diet can cause damage to intestinal mucosa, inhibit protein absorption and lead to intestinal epithelial hyperplasia (Ma et al., 2012). Undigested proteins and amino acids are transported to the hindgut as substrates for pathogenic fermentation bacteria, causing diarrhea. Therefore, reasonable protein intake is very important to human health.
In addition, more and more studies have proved that the flora is involved in protein metabolism and the balance adjustment of amino acids, and the flora is related to some diseases caused by protein metabolism disorders (Xu et al., 2015), so the regulation of the intestinal flora can be used for clinical medicine.
2.3. Phenolics
Phenolic compounds are secondary metabolites (Chambers et al., 2015) of plants. There are thousands of phenolic compounds and their derivatives. Polymerization and glycosylation further increase the complexity and diversity of these compounds. They are usually called polyphenols (Tsuda, 2016). All plants produce polyphenols, and most of them exist in glycosylated form (Cardona et al., 2013). A number of studies have shown that polyphenols have anti-cancer, anti-inflammatory, and anti-oxidative effects, and have a two-way regulatory effect on the intestinal flora. Polyphenols will be recognized by the human body as heterologous organisms after being ingested, so their bioavailability is relatively low compared with trace and macronutrients. Some simple low molecular weight polyphenols can be absorbed in the small intestine (Appeldoorn et al., 2009). During the absorption process, polyphenols are usually hydrolyzed by intestinal enzymes, and then are absorbed in the intestine and liver cells through methylation, sulfation or glucuronidation (Del Rio et al., 2010). As a result, a series of water-soluble conjugated metabolites (methyl, glucuronic acid and sulfate derivatives) are quickly released to the systemic circulation for further distribution to organs and urine excrement (Cardona et al., 2013). But the small intestine can only absorb 5–10% of the total polyphenol intake. The remaining polyphenols (accounting for 90–95% of total polyphenol intake) may accumulate in the lumen of the large intestine until the millimolar range, plus the conjugates are excreted into the intestinal lumen through bile, they will be affected by the enzymes of the intestinal microorganism. The promoting effect of dietary phytochemicals is transformed into absorbable metabolites (Rothwell et al., 2012). Therefore, in addition to individual differences in the daily intake of polyphenols, individual differences in the composition of the intestinal flora may also lead to differences in the bioavailability and biological efficacy of polyphenols and their metabolites (Gross et al., 2010).
The interaction between polyphenols and microbiota has a dual nature. Different polyphenols and their metabolites have different effects on the intestinal flora, such as tea polyphenols on Clostridium perfringens, Staphylococcus aureus, and Vibrio parahaemolyticus. Due to the inhibitory effect, and the methanol extract of green tea can selectively enhance the growth of Bifidobacteria (Hara et al., 1995). The mechanism of action of polyphenols on the intestinal flora is very complicated, but it mainly affects the number and types of intestinal flora. As the number and types of intestinal flora change, its function will also change. The effect of polyphenols on bacterial growth and metabolism depends on the structure of polyphenols, the measured dose and microbial strains, which mainly affect microbes through the following mechanisms (Hervert-Hernandez and Goni, 2011). One is to inhibit the growth of microorganisms by destroying the function of cell membranes (Kemperman et al., 2010). Polyphenols can block the electron chain transmission and oxidative phosphorylation of the cell membrane and bind to the bacterial cell membrane in a dose-dependent manner, destroying membrane function. Polyphenols (such as catechins) also act on different bacterial species by producing hydrogen peroxide and changing its permeability (Haslam et al., 1992). In addition, the influence of polyphenols on microorganisms is also related to the self-defense mechanism of microorganisms. Microorganisms exposed to polyphenols up-regulate proteins related to defense mechanisms, thereby protecting cells, while down-regulating various metabolic and biosynthetic proteins involved in amino acid and protein synthesis, as well as phospholipid, carbon, and energy metabolism (Xuelian, 2014). Most bacteria can adjust phenotypic characteristics, including virulence factors, according to cell density under the control of chemical signaling molecules (González and Keshavan, 2006). Polyphenols also interfere with bacterial quorum sensing by producing and releasing small signal molecule compounds that are self-inducers, such as acylated homoserine lactones in Gram-negative bacteria and oligopeptides in Gram-positive bacteria (Huber et al., 2003). According to reports, polyphenols can interfere with the production of small signal molecules in bacterial cells of Escherichia coli, Pseudomonas putida, and Burkholderia cepacia, which triggers the exponential growth of bacterial populations (Smith et al., 2005). Polyphenols also have the property of chelating with metal ions to synthesize insoluble complexes, so they can deprive metal ions (such as iron, cobalt, etc.) of metal enzymes in intestinal microbial cells, thereby inhibiting enzyme activity, and affecting aerobic microorganisms that require iron to function (Freestone et al., 2007). On the contrary, studies have shown that catechol can provide iron to promote the growth of specific intestinal bacteria in the case of iron deficiency (Kim et al., 2018). It further proves the duality of polyphenols in mediating the intestinal flora, but its complicated influence mechanism still needs further research.
| 3. Effect of dietary phytochemicals on intestinal flora-related diseases | ▴Top |
3.1. IBD
Enteritis is closely related to intestinal microecology. It was found that there was obvious imbalance of intestinal flora in patients with enteritis (Pascal et al., 2017), as shown in Figure 3. Bacteroides and scleroderma were significantly reduced in the intestinal tract of IBD patients, while the harmful bacteria such as Actinomycetes and Proteus increased (Kostic et al., 2014). Compared with healthy people, the intestinal microflora of CD patients contains a large number of adhesive infiltrating Escherichia coli, pathogenic Yersinia and Clostridium difficile (Zhilu et al., 2019).
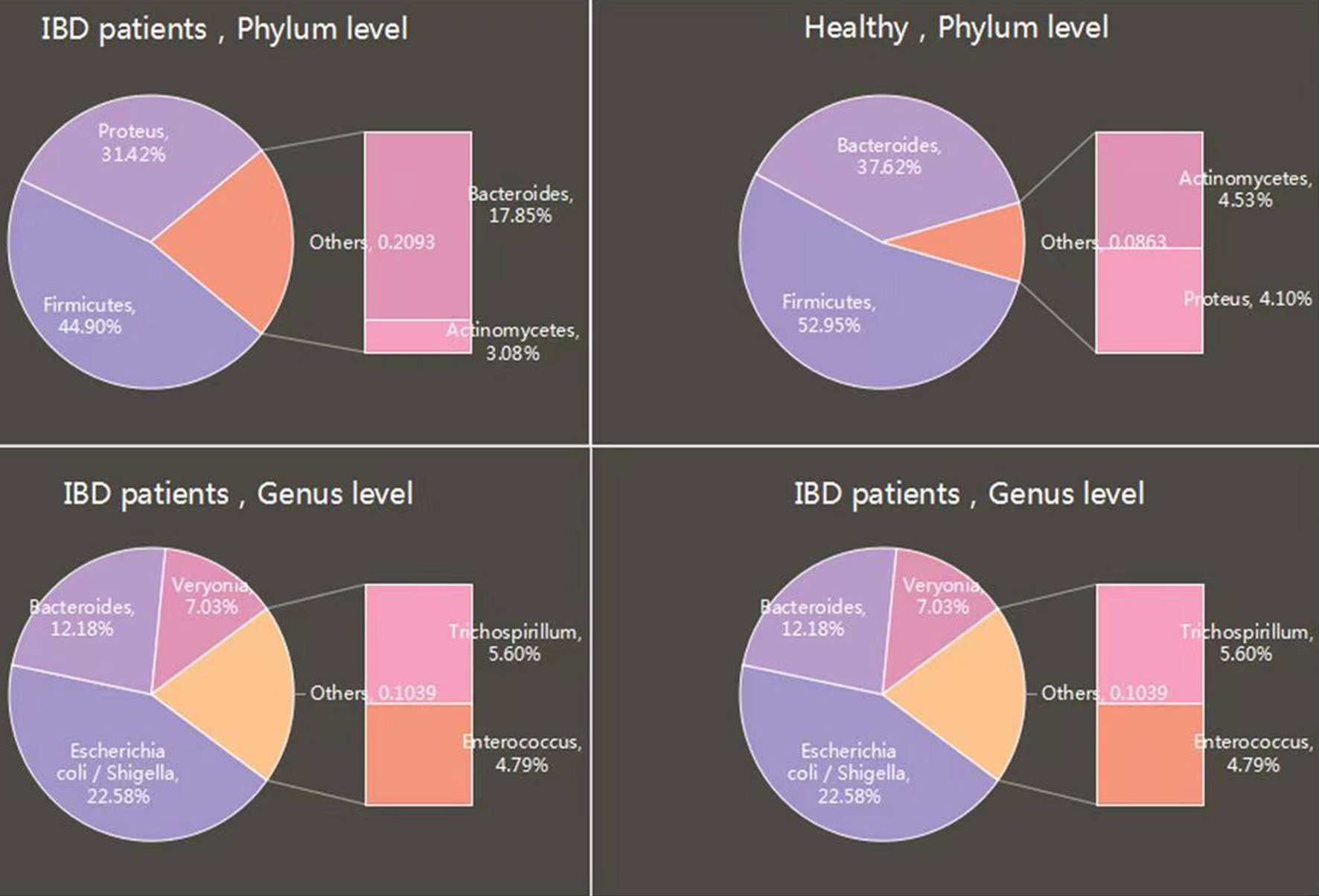 Click for large image | Figure 3. Comparison of intestinal microbial diversity between IBD patients and healthy people. |
Intestinal flora plays an important role in intestinal homeostasis from three aspects. The gut microbiota supplies energy and nutrients to the host (O’Hara and Shanahan, 2006). As mentioned earlier, intestinal bacteria also provide SCFAs by fermenting resistant starch or indigestible carbohydrates (dietary fiber). The changes of intestinal microflora can reduce the metabolism of SCFA, reduce the synthesis of biosynthetic amino acids, and lead to nutritional defects, oxidative stress and endotoxin secretion (LeBlanc et al., 2017). This may be a key factor that compromises intestinal and immune homeostasis (Machiels et al., 2014).
Intestinal flora plays an important role in intestinal homeostasis by affecting the host immune system. Mice lacking intestinal microbiota showed immature lymphoid tissue (Bouskra et al., 2008), decreased number of intestinal lymphocytes, and decreased levels of antimicrobial peptides (Cash et al., 2006) and immunoglobulin Ig A (Hapfelmeier et al., 2010). These defects and abnormalities of the immune system can be restored by intestinal microbiota colonization.
The gut microbiota contributes to host defense against pathogens. The gut microbiota enhances colonization resistance to intestinal pathogens by both direct and indirect mechanisms of action. Some commensal bacteria directly inhibit intestinal pathogens by competing for nutrients or by inducing the production of inhibitory substances. For example, Bacteroides (B.) thetaiotaomicron, which is an abundant colonic anaerobe, consumes carbohydrates used by C. rodentium, which contributes to the competitive exclusion of the pathogens from the intestinal lumen (Cardona et al., 2013). In conclusion, dietary habit and intestinal flora are closely related to IBD.
3.2. Obesity
As one of the mechanisms of intestinal inflammation, low-grade obesity has attracted more and more attention (Tsukumo et al., 2009). (a) Intestinal microbes can metabolize and utilize the hard to digest food ingredients of human body to obtain energy. (b) Intestinal flora regulates the increase of plasma lipopolysaccharide levels, thus increasing the incidence of inflammation, obesity and diabetes. (c) gut microbes induce host gene regulation that regulates energy consumption and storage patterns. The results showed that the number and diversity of intestinal microflora in obese patients were significantly decreased, which was mainly manifested by the significant reduction of Clostridium perfringens and Bacteroides, and the increase in the number of Firmicum bacteria (Sharma et al., 2020). At the genus level, the abundance of SCFA producing Lactobacillus was higher in intestinal contents, and the relative abundance of Gram-negative bacteria such as Helicobacter pylori, Sphingomonas and Vibrio desulphurizing in colonic mucosa was higher (Kim et al., 2015). Al-Attas and other studies found that Bacteroides decreased by about 50% in obese mice, while Firmicum increased by the same extent (Al-Attas et al., 2009). Alissa Nicolucci et al. found that fructooligosaccharide enriched inulin can selectively change the intestinal flora, significantly reduce the weight of overweight or obese children, increase the species richness of Bifidobacteria, and reduce the number of vulvar Bacteroides (Nicolucci et al., 2017). Another study has shown that intestinal microbiota can ferment indigestible carbohydrates into SCFAs, which are important for maintaining energy homeostasis (Cummings, 1981). Fernandes et al. found that the level of SCFAs in feces of obese individuals was significantly higher than that of lean people, indicating that the increased level of SCFAs in feces is closely related to the occurrence of obesity (Fernandes et al., 2014). Recently, a new intestinal microbiota has been found to improve obesity, and Christensen has been shown to regulate the composition of intestinal microbiota and reduce the weight of mice (Conterno et al., 2011). After administration of inulin, the number of Clostridium perfringens and Eubacterium rectum increased (Hippe et al., 2011). Both of them are butyric acid producing bacteria. Butyric acid has anti-inflammatory, anti-cancer, improving the integrity of intestinal barrier, and can prevent the development of metabolic disorders (Qin et al., 2010). In addition, dietary habits are closely related to intestinal microflora. High fat diet can lead to changes in intestinal microbial spectrum and decrease in diversity (An et al., 2018). Bacteroides showed a downward trend, while Clostridium showed an upward trend (Carmody et al.,2015). High carbohydrate intake resulted in increased gene abundance of intestinal flora (Koliada et al., 2017), increased levels of beneficial Bifidobacteria (Pokusaeva et al., 2011), and decreased growth of opportunistic pathogens such as Mycobacterium avium, subspecies Paratuberculosis and Enterobacteriaceae (Walker et al., 2011). Therefore, healthy eating habits are very important.
| 4. Conclusion and future perspectives | ▴Top |
The intestinal microbiota in human body can be divided into two types: beneficial and harmful. Bifidobacterium, Lactobacillus and other beneficial bacteria account for more than 90% of the total, as shown in Figure 4. With the understanding of this relationship, it has been found that the intestinal microbial community is closely related to the biological antagonism, immunity, nutrition, tumor and acute and chronic infection of the host. It participates in various physiological and pathological processes and is an important part of human body. Therefore, it is worth exploring how to regulate the intestinal microflora to improve the health status and enhance the potential disease resistance of animals. In general, they can increase beneficial bacteria and reduce harmful bacteria. In this sense, they have considerable utilization value and economic benefits. It can provide new ideas for the treatment and prevention of many diseases.
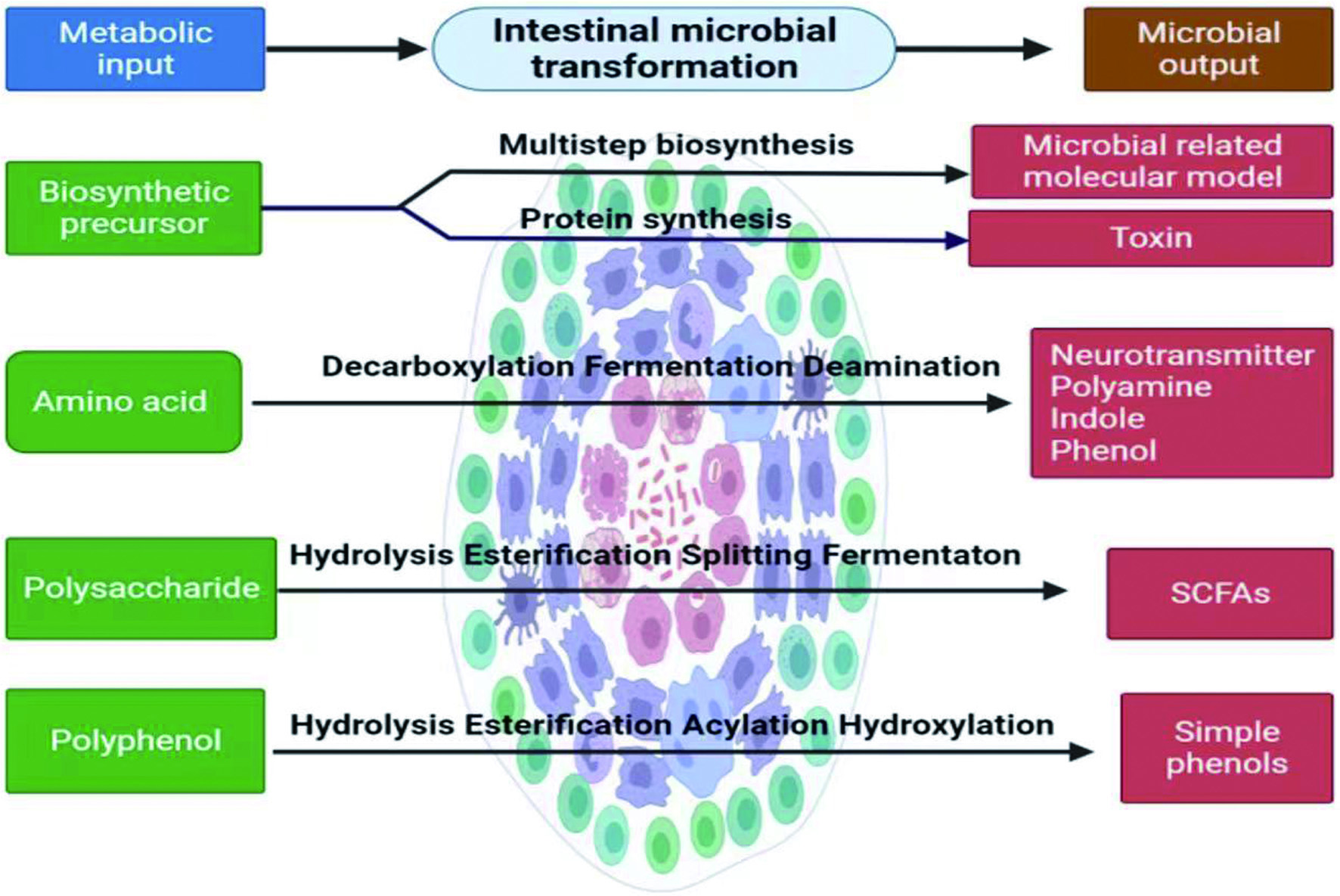 Click for large image | Figure 4. Intestinal flora transformation. This figure was created with icons provided by bio render (https://biorender.com). |
Insights into how plant compositions impact the gastrointestinal microbiota are emerging. However, more research is needed to determine whether the regulation of plant components on intestinal flora can be translated into benefits for human health. Large prospective studies are necessary to determine the directionality of the associations between perturbations in the microbiota and disease. Through animal experiments, isotope labeling and high-throughput analysis techniques can be used to further determine the transport pathway of plant components in vivo and their impact on intestinal flora. Moreover, microbial metabolites and other physiological measures of health such as blood cholesterol, glycemia, and inflammation, also need to be evaluated. Most of these efforts have so far been directed towards in vitro studies, and for these reasons the clinical gap is still substantial. When possible, well-controlled clinical trials can be carried out. Besides, in this era of rapid technological and computational advances, efforts should be made to move beyond simple characterization of the composition of the microbiota and toward functional activities of the microbiota through transcriptomics, metabolomics, and proteomics. Through integrate vast multi-omics data sets allow us to extend our understanding of host-microbe interactions.
Acknowledgments
This work was kindly funded by the Natural Science Foundations of Hebei Province (C2019407092), Hebei province’s key research and development program (216Z3201G).
These authors contributed equally to this study.
Conflict of interest
The authors declare no conflict of interest.
Bin Du: Investigation; Writing. Zilong Ma: data curation; Writing-original draft. Guang Xin: Review & editing. Yuedong Yang: Conceptualization. Baojun Xu: Conceptualization; Data curation; Supervision; Editing.
| References | ▴Top |