| Journal of Food Bioactives, ISSN 2637-8752 print, 2637-8779 online |
| Journal website www.isnff-jfb.com |
Review
Volume 15, September 2021, pages 21-28
Phytochemicals: Potential Lead Compounds for COVID-19 Therapeutics
Srishti Kashyapa, Revathy Nadhana, Danny N. Dhanasekarana, b, *
aStephenson Cancer Center, The University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, USA
bDepartment of Cell Biology, The University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, USA
*Corresponding author: Danny N. Dhanasekaran, Stephenson Cancer Center, The University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, USA. E-mail: danny-dhanasekaran@ouhsc.edu
DOI: 10.31665/JFB.2021.15279
Received: September 27, 2021
Revised received & accepted: September 29, 2021
| Abstract | ▴Top |
Coronavirus Disease 2019 (COVID-19) is a global pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS- CoV-2). The rising number of cases of this highly transmissible infection has pressed for the urgent need to find effective therapeutics. The life cycle of SARS-CoV-2 includes the viral entry, viral replication, viral assembly and release. The symptoms associated with viral infection often leads to fatal outcome with pneumonia, myocarditis, acute respiratory distress syndrome, hypercoagulability, and/or multi-organ failure. Recent studies have reported that phytochemicals such as emodin, epigallocatechin gallate, and berberine could, albeit modestly, inhibit different stages of SARS-CoV-2 life cycle. The phytochemicals have been shown to disrupt viral infection and replication by blocking viral-surface spike protein binding to entry receptor angiotensin-converting enzyme (ACE2), inhibiting viral membrane fusion with host cells, inhibiting RNA-dependent RNA polymerase involved in viral replication, and/or pathological host- responses in vitro. The focus of this review is to evaluate the efficacies of these phytochemicals on inhibiting SARS-CoV-2 viral infection, growth, or disease progression as well as to provide a perspective on the potential use of these phytochemicals in the development of novel therapeutics against SARS-CoV-2.
Keywords: SARS-CoV-2; COVID-19; Coronavirus; Phytochemicals; Cytokine Storm; Cytokine Release Syndrome
| 1. Introduction | ▴Top |
The worldwide outbreak of highly transmissible Coronavirus Disease-2019 (COVID-19) was caused by a zoonotic pathogenic virus, Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). SARS-CoV-2 and its previous counterpart, SARS-CoV, which resulted in a pandemic in 2002, belong to the β-coronavirus lineage B, both of which are similar to the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) that had emerged in 2012 (Zaki et al., 2012). Towards the end of 2019, the emergence of the COVID-19 pandemic posed an unprecedented global health threat, prompting the need to develop effective antiviral therapeutics immediately. During the initial stage of the pandemic, repurposing of the existing drugs was considered to be one of the most tangible options for rapidly developing the effective therapeutics. Numerous clinical trials have been completed, but none of those evaluated repurposed drugs could give any promising results (Martinez & chemotherapy, 2020). The failure of these repurposed drugs in their clinical trials stresses the need to unleash novel structural motifs for developing effective COVID-19 drugs. A historical overview of drug discovery has shown that natural products could be well optimized for efficacious therapeutic strategies for many diseases including infectious diseases. The uniqueness of the vast array of chemical structures of natural products has been the inspiration behind the development of several effective pharmaceuticals (Clercq, 2007). In this review, we briefly discuss the potential of several plant-derived natural products or phytochemicals, which could serve as lead compounds for synthesizing effective nature-inspired therapeutics against COVID-19.
| 2. COVID-19 virus and its life cycle | ▴Top |
SARS-CoV-2 genome has a length of nearly 30,000 nucleotides and is the largest positive-sense genome of the RNA virus family (Astuti and Ysrafil, 2020). The genetic material of the virus contain a simple structure consisting of a single stranded RNA genome and bound ribonucleoprotein particles that provide structural integrity to RNA in addition to playing a role in viral replication. This RNA-RNP complex is encapsulated in a viral envelope consisting of lipid membrane. The viral envelope is interspersed with envelope and membrane proteins. The outward projecting and the receptor-interacting spike glycoproteins are also embedded in the viral envelope (Figure 1). The infection cycle of the virus can be divided into three stages, namely, viral entry, viral RNA release and replication, and viral assembly and release. To enter the host cells, coronaviruses first bind to specific host cell surface receptors and then the viral Spike protein is cleaved by host cell-derived proteases to enable fusion of viral and cellular membranes (V’kovski et al., 2021). Receptor binding domain (RBD) of SARS-CoV-2 Spike (S) protein specifically recognizes human angiotensin- converting enzyme-2 (ACE-2) as its receptor (Letko et al., 2020; Wan et al., 2020). Proteolytic cleavage of S protein by host cell surface protease, TMPRSS2, leads to the final conformational changes in the S2 domain of the spike protein needed for virus-host cell membrane fusion. This process is irreversible and it is stringently regulated (Walls et al., 2017). The release of SARS-CoV-2 viral genome in the host cell cytoplasm marks the onset of spatially and temporally regulated complex program of viral gene expression.
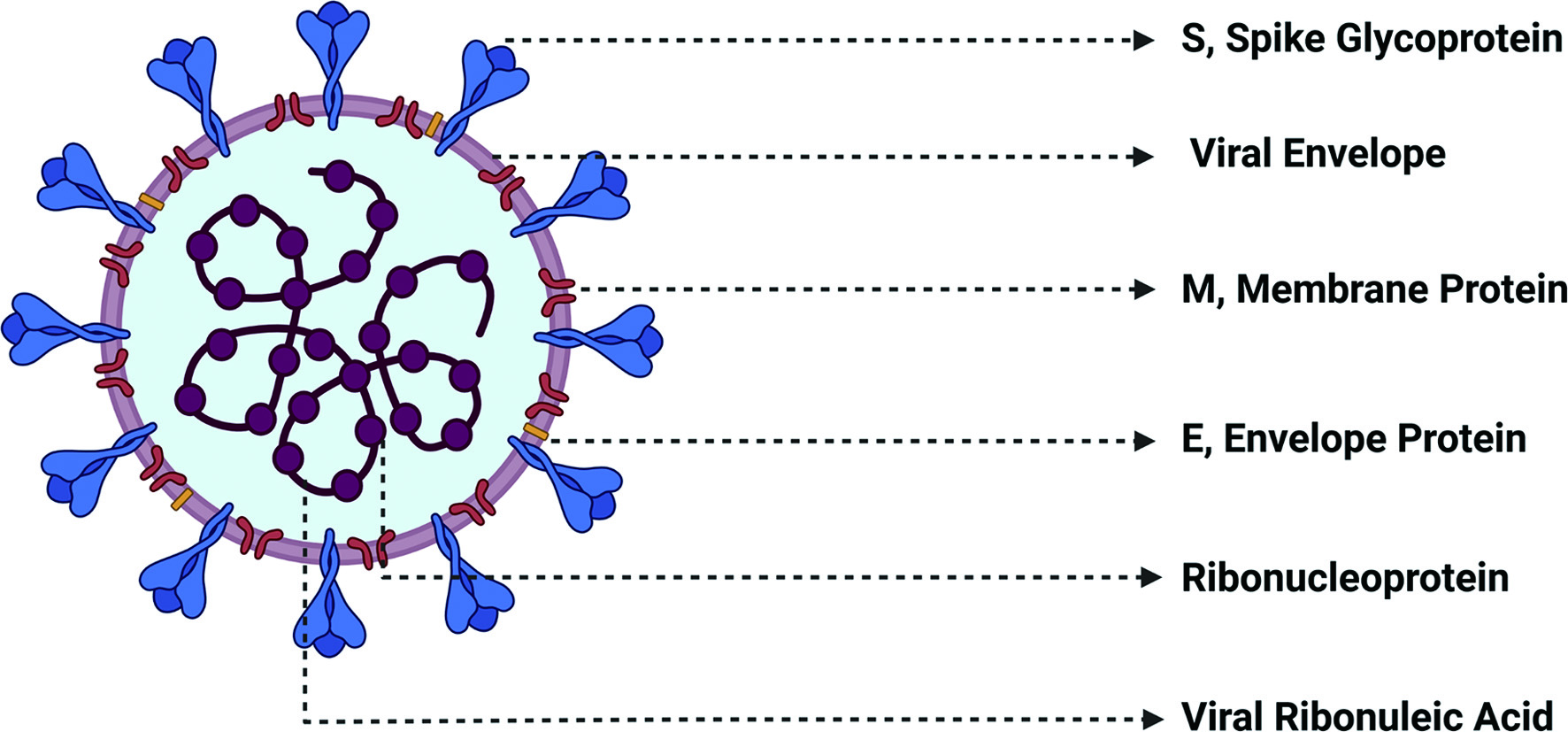 Click for large image | Figure 1. Structure of SARS-CoV-2 Virus. SARS-CoV-2 virus contains a single stranded RNA serves as the genetic material to which ribonucleoproteins that assist in viral structure and replication.are bound. The encapsulating viral envelope is comprised of a lipid layer interspersed with membrane and envelope proteins as well as characteristic receptor-interacting spike glycoprotein. |
Coronavirus particles contain a single, 5′-capped and 3′-polyadenylated RNA genome which codes for two large overlapping open reading frames in gene 1, as well as a variety of structural and nonstructural proteins at the 3′ end. At the 5′ end, SARS-CoV-2 genomic RNA comprises of two overlapping reading frames ORF1a and ORF1b (Gorbalenya et al., 2006). The translation of ORF1a and ORF1b from the genomic RNA produces two polyproteins, pp1a and pp1ab, respectively. The pp1a and pp1ab proteins are then cleaved by the viral proteases, papain-like protease (PLPRO) and 3C-like protease (3CLPRO) to release 16 different non-structural proteins (Nsp). These encoded Nsp primarily comprises of RNA-dependent RNA polymerase (RdRp), helicase, and exonuclease (ExoN), which together forms a viral replication and transcription complex (RTC) (Subissi et al., 2014). The RdRp contains a catalytic subunit, NSP12, helicase (NSP13), and accessory subunits NSP7 and NSP8 (Hillen et al., 2020). The RdRp complex transcribes the viral genome template of negative-sense genes, including progeny genome and subgenomic RNA as intermediate products, followed by the transcription of positive sense mRNAs mediated by RdRp. ExoN, exclusive to the β-coronavirus family, conducts RNA proofreading activity, which is critical for maintaining the viral genome integrity. The 3′ end of the viral genome contains the ORFs which encode for viral structural proteins, namely the spike (S), the membrane (M), the envelope (E), and the nucleocapsid (N) proteins, in addition to the accessory proteins. These structural proteins would be incorporated into the endoplasmic reticulum membrane and forms the membrane encased structures, which further encloses the nucleocapsid and other necessary accessory proteins to form the exocytic vesicle. The exocytic vesicle fuses with the plasma membrane and releases the viruses into the extracellular space, which spreads throughout the human body (Murgolo et al., 2021; Romano et al., 2020; Trougakos et al., 2021). This could elicit pathological inflammatory response including cytokine release syndrome and cytokine storm. Eventually, multiple organ failure ensues with the resultant death of the COVID-19 patient.
In spite of the fact that numerous drugs do exist that have clinically proven antiviral properties (Indari et al., 2021) and several drugs that show a limited potential to alleviate COVID-19 symptoms (Lam et al., 2020; Raju et al., 2021), none of them have proven to work efficiently to reduce COVID-19 associated mortality. Nevertheless, several in vitro studies have demonstrated the potential of several phytochemicals to inhibit COVID-19 infectious cycle in humans at various stages (Alam et al., 2021; Behl et al., 2021; Marmitt et al., 2021; Merarchi et al., 2021). These phytochemicals have been demonstrated to inhibit viral entry, viral replication, or viral release – in addition to attenuating the host pathological responses (Figure 2). However, these studies have fallen short of proving a preventive or therapeutic role for these phytochemicals against COVID19 infection in real-time. Nevertheless, they provide strong evidence that they can serve as potential lead molecules for either the therapy or the prevention of SARS-CoV-2 infection as well as other viral infections that might emerge in the future (Swain et al., 2021).
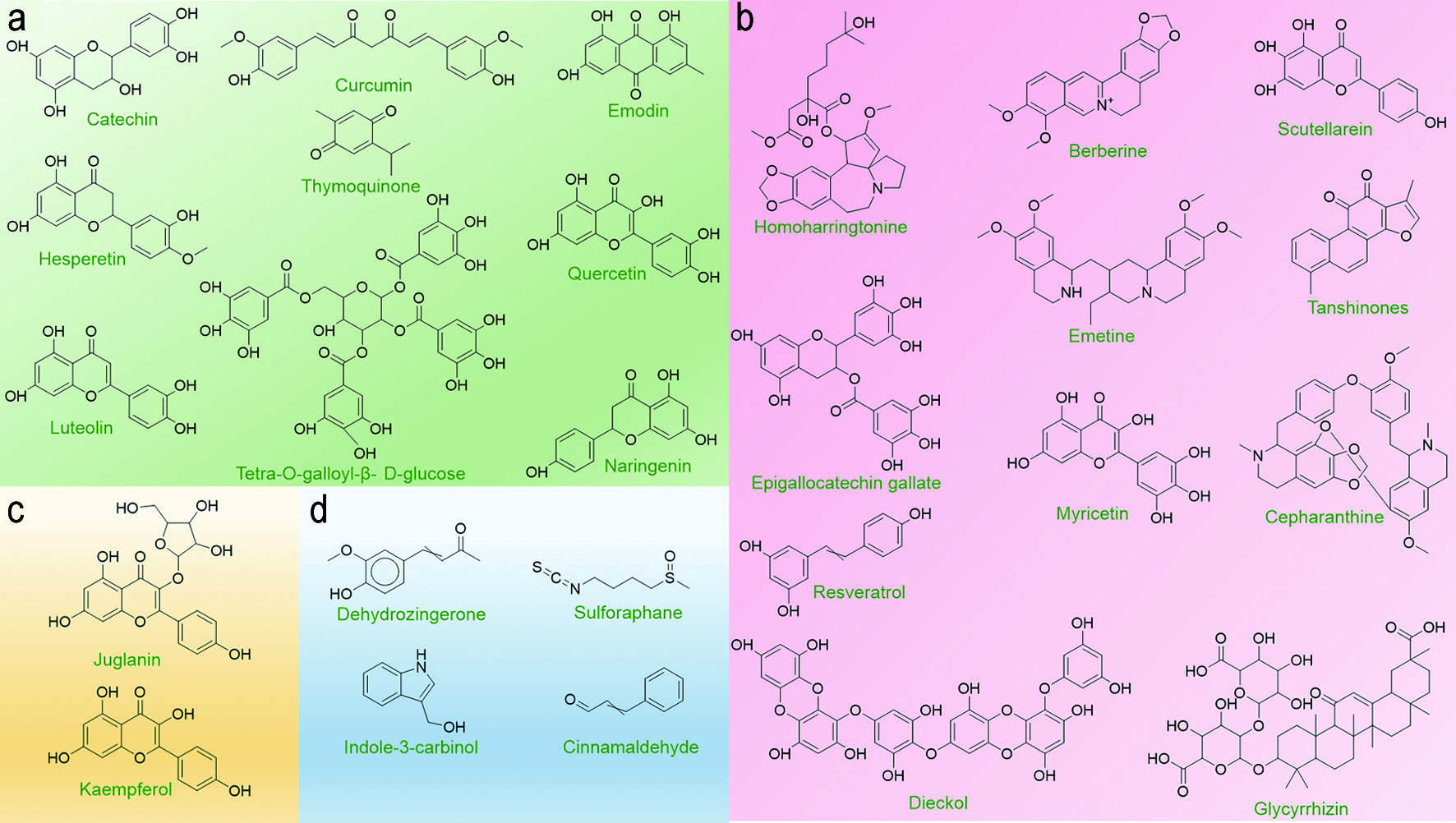 Click for large image | Figure 2. Potential Lead Phytochemicals that can target SARS-CoV-2. Several phytochemicals have been identified to have the potential to disrupt different stages of viral lifecycle or pathological host response following infection. The structures of candidate phytochemicals that have potential to serve as the lead compounds for drug discovery are presented as follows: (a) Phytochemicals targeting viral entry into the host cell; (b) Phytochemicals targeting viral replication; (c) Phytochemicals inhibiting viral release from the host cell; and (d) Phytochemicals that can attenuate host CRS or cytokine storm (See Text for Details). |
| 3. Phytochemicals targeting SARS-CoV-2 entry into the host cell | ▴Top |
Coronavirus entry into the host cell is one of the most important determinant of viral infectivity (Li, 2016) and is also the major target for human intervention strategies (Du et al., 2009b) . Coronavirus spike (S) proteins are homotrimeric class I fusion glycoproteins that comprise of two functionally distinct subunits: S1 and S2. The peripheral S1 subunit contains the receptor-binding domain that binds to the host cell receptor, ACE2 and is critical in determining tropism of infection and pathogenicity (Li et al., 2003; Zhou et al., 2020). The transmembrane S2 subunit that contains the fusion peptide domain and the twin heptad repeat domains mediates viral and cellular membrane fusion upon extensive conformational rearrangements following host cell receptor binding (Letko et al., 2020). The S protein is a class I viral fusion membrane glycoprotein similar to HIV glycoprotein 160 (Env), influenza haemagglutinin (HA), and Ebola virus glycoprotein (Du et al., 2009). In the first step of viral entry, the peripheral amino (S1) subunit of the SARS-CoV-2 spike glycoprotein (S) independently binds with the membrane metallopeptidase ACE2, and the carboxy (S2) terminus embedded into the viral envelope mediates fusion of viral and cellular membranes (Gallagher and Buchmeier, 2001). Inhibiting the first step of viral infection significantly minimizes the chance for the virus to propagate, evolve, and eventually acquire drug resistance (Breitinger et al., 2021; Esté, 2003; Horne and Vohl, 2020). Therefore, several phytochemicals have been investigated for their ability to exhibit inhibitory effect on viral entry (Figure 3).
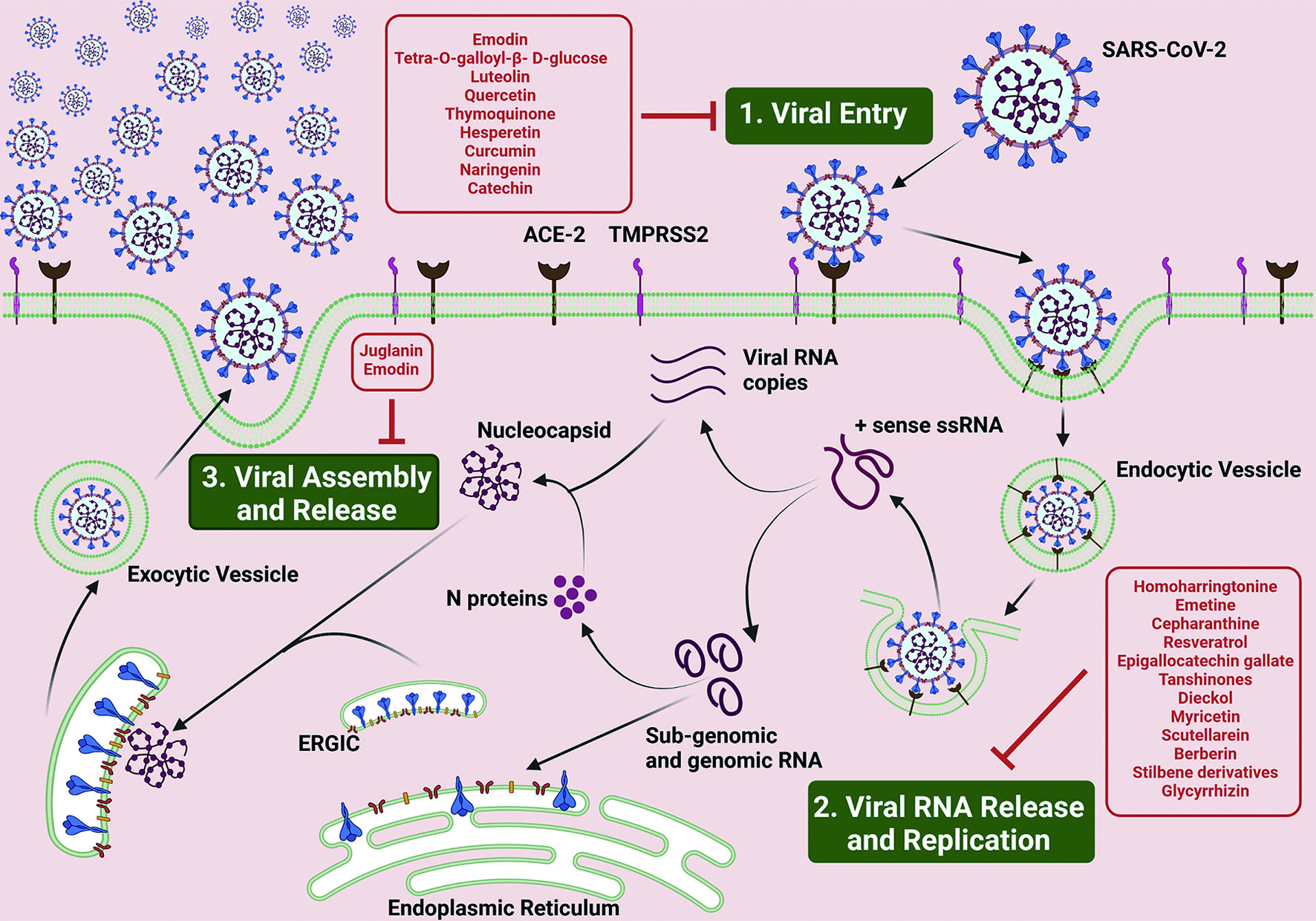 Click for large image | Figure 3. Lead Phytochemicals that can target SARS-CoV-2 Life Cycle. SARS-CoV-2 life cycle in human host is broadly divided in to three steps, namely, 1. Viral Entry; 2. Viral RNA Release and Replication; and 3. Viral Assembly and Release (see text for details). Briefly, SARS-CoV-2 viruses attach to the ACE-2 receptor on the plasma membrane of the host cell via the interaction of S, Spike glycoprotein. This interaction triggers the activity of TMPRSS2 receptors present on the plasma membrane, initiating the endocytosis of the viral particles. Upon entry, the viral particle encapsulated in the plasma membrane structure, the endocytic vesicle, opens up to release the viral RNA into the host cell cytoplasm. The ss RNA undergoes replication to produce numerous RNA viral RNA replicates as well as genomic and sub-genomic RNA, the former which fuses with N proteins to form the nucleocapsid structure and the latter which translates into the S, M and E envelope proteins and the accessory proteins. The membrane proteins incorporates into the endoplasmic reticular membrane which releases off into the cytoplasm and encloses the nucleocapsid structures to form exocytic vesicle, which then fuses with the host cell plasma membrane and releases the viruses into the extracellular space. Phytochemicals with the potential to disrupt different stages of the viral infection cycle are boxed and listed in red. |
Emodin, an anthraquinone polyphenol obtained from traditional herbs Rhubarb (Rheum officinale) and Japanese knotweed (Polygonum cuspidatum), significantly inhibited the interaction of S protein with ACE2 in a dose-dependent manner, in a cell-free competition assay, with an IC50 of 200 μM (Ho et al., 2007). Emodin also reduced the infection of ACE2-expressing Vero E6 cells, a non-human primate kidney cell-line, by an S protein-pseudo typed retrovirus (Ho et al., 2007). High-throughput screening of a library containing the extracts from 121 Chinese herbs led to the identification of two candidate low molecular weight phytochemicals, namely Tetra-O-galloyl-β- D-glucose (TGG) and luteolin (Yi et al., 2004). TGG and luteolin inhibited SARS-CoV infection by preventing viral entry into Vero E6 cells with the EC50 values of 4.5 μM and 10.6 μM, respectively. Frontal affinity chromatography-mass spectrometry studies revealed that both TGG and luteolin bound with high affinity to SARS-CoV spike protein, suggesting that the inhibition of viral entry could be due to the interference in the interaction between ACE2 and spike protein (Yi et al., 2004). TGG is a polyphenolic compound obtained from Indian gooseberry (Phyllanthus emblica), and Luteolin is a common flavonoid found in vegetables and fruits such as celery, parsley, broccoli, onion leaves, carrots, peppers, cabbages, and apple skin (Miean and Mohamed, 2001). Molecular docking and SPR studies have revealed that thymoquinone isolated from black cumin (Nigella sativa)(Harzallah et al., 2011) binds to ACE2 with high affinity, with a Kd value of 32.140 μM. Thymoquinone inhibited the infectivity of SARS-CoV-2 pseudovirus in HEK293T-ACE2 in a dose-dependent manner with an IC50 of 4.999 μM (Xu et al., 2021).
Molecular docking and dynamic simulation studies predict polyphenols from Citrus sp. (hesperetin, hesperidin, and tangeretin) and Curcuma species (curcumin and its derivatives) to have a potential inhibitory effect on SARS-CoV-2 infection by interacting with the S protein RBD (Utomo & Meiyanto, 2020). Hesperidin was predicted to dock at the binding interface between S protein and ACE2 by positioning on the shallow middle pit of the surface of the S protein RBD (Wu et al., 2020). Naringenin, a flavanone found in citrus fruits (grapefruit, sour orange), tomato and other fruits, was reported to have stronger binding energy with the spike glycoprotein than remdesivir (Ubani et al., 2020), an anti-viral drug temporarily approved by the FDA in the treatment of COVID-19 (Hendaus & Dynamics, 2020). Being SARS-CoV-2’s point of entry into the host cells, ACE2 has also been interrogated as a potential target for anti-viral drug discovery. Computational studies showed that catechin and curcumin exhibited strong binding affinity not only to the viral Spike protein and the host receptor ACE2, but also their complex (Spike-RBD/ACE2-complex), with binding affinity values of −10.5 and −7.9 kcal/mol; −8.9 and −7.8 kcal/mol; and −9.1 and −7.6 kcal/mol, respectively (Jena et al., 2021). By interfering with the host cell receptor and viral spike protein interaction, catechin and curcumin could have an inhibitory effect on the cellular entry of the virus.
| 4. Phytochemicals targeting SARS-CoV-2 replication within the host cell | ▴Top |
Natural compounds which inhibit viral proteases can be viewed as potential candidates for managing COVID-19 (Figure 3). Protease inhibitors have been used to treat and manage viruses that cause HIV-AIDS, MERS, and SARS (Kumar et al., 2020). Replication of SARS-CoV-2 requires proteolytic processing of the replicated polyproteins by viral proteases leading to the release of non-structural and structural proteins. Tanshinones derived from Salvia miltiorrhiza (red sage) moderately inhibit the SAR-CoV 3CLpro and PLpro proteases at IC50 values ranging from 0.8 to 30 μM (Park et al., 2012).
Dieckol, a pholorotannin obtained from Ecklonia cava, is reported to exert significant inhibitory activity (IC50= 2.7 μM) on coronavirus 3CLpro. Dieckol which possess two eckol groups linked by a diphenyl ether, also exhibited potent inhibitory activity on the cell-based 3CLpro cis-cleavage assay, and its potency was greater than the other phlorotannin derivatives and the natural reference inhibitors (Park et al., 2013). Myricetin, a flavonoid commonly present in nuts, berries, tea, wine, and Scutellarein, a flavone, inhibits the SARS-CoV helicase protein in vitro by affecting its ATPase activity, but not the unwinding activity (Yu et al., 2012).
Recent studies showed that homoharringtonine (HHT) and emetine inhibited SARS-CoV-2 replication in Vero-E6 cells with EC50 at 2.55 and 0.46 μM, respectively (Choy et al., 2020). Lycorine, emetine, and cephaeline may inhibit viral protein synthesis by interacting with the host ribosome (Ren et al., 2021). Berberine an alkaloid isolated from plants such as Berberis has been reported to inhibit the replication of SARS-CoV-2 in cultured nasal cell lines (Varghese et al., 2021). Resveratrol, a well-known polyphenol inhibits SARS-CoV-2 viral replication (Pasquereau et al., 2021; ter Ellen et al., 2021; Yang et al., 2021). Glycorhyzin, a triterpene saponin, found in high concentrations in the root of the Glycyrrhiza glabra (liquorice) plant inhibits viral replication in vitro by inhibiting the CLpro protease (van de Sand et al., 2021). EGCG, a major component in green tea, has also been shown to inhibit viral replication in Vero cells. This inhibition is associated with EGCG’s ability to inhibit Nsp15, a non-structural protein involved in the processing of the viral genome for replication (Hong et al., 2021). In addition to the thymoquinones which has been reported to inhibit the viral entry into the host cells, plumbagin, which is a naphthoquinone, isolated from roots of Plumbago zeylanica has been predicted to induce viral RNA degradation, through its ROS inductive effects, thereby exerting an antiviral therapeutic response (Nadhan et al., 2021).
| 5. Phytochemicals targeting SARS-CoV-2 viral release from the infected host cell | ▴Top |
Phytochemicals belonging to Kaempferol family of flavanol such as Juglanin has been tested for their ability to inhibit the release of assembled mature virions from the host cell into extracellular space to propagate cell-to-cell spread of the infection (Figure 3). The ORF 3a of SARS-CoV codes for a transmembrane protein that forms a homotetrameric ion channel and modulates viral release (Lu et al., 2006). It can be reasoned that the phytochemicals that inhibit the ion channel could exert an inhibitory effect on virus release, and they could serve as lead compounds for the development of novel therapeutic antiviral agents. Kaempferols are natural flavonols, found in plants such as kale, beans, tea, spinach and broccoli (Baenas et al., 2012). Several kaempferol derivatives were reported to moderately block the 3a channel of Coronavirus, hence, inhibiting the viral production and release from the host cells (Schwarz et al., 2014). Kaempferol glycoside derivatives were proved to possess more potent inhibitory effect than kaempferol and points towards the significance of sugar residues for the antiviral activity (Schwarz et al., 2014) Juglanin (kaempferol-3-O-a-L-arabinofuranoside) was the most effective glycoside with an IC50 value of 2.3 μM. Other kaempferol glycosides i.e. tiliroside (kaempferol-3-O-(6-p-coumaroyl)-glucoside) and afzelin (kaempferol-3-O-a-L-rhamnoside) were less potent than juglanin, but showed similar activity to that of emodin (Schwarz et al., 2014).
| 6. Phytochemicals targeting cytokine release syndrome/cytokine storm | ▴Top |
In addition to this array of phytochemicals that target viral entry, replication, and release, there has been increased focus on phytochemicals that can have modulatory effect on Cytokine Release Syndrome (CRS) and cytokine storm of the infected patients (Figure 4). CRS and cytokine storm can be broadly described as life-threatening systemic inflammatory syndromes. In CRS and cytokine storm, elevated levels of inflammatory cytokines and excessive activation of immune cells lead to a range of clinical manifestations such as fever, fatigue, disseminated intravascular coagulation (DIC), hypotension, hemostatic imbalance, respiratory failure (acute respiratory distress syndrome - ARDS), multi-organ failure, and even death, if not treated in time (Mangalmurti and Hunter, 2020). CRS and cytokine storm are primarily associated with infectious and immune-mediated conditions. Severe pneumonia with elevated cytokine levels such as interferon-γ (IFNγ), Interleukin (IL)- 6, and granulocyte-macrophage colony stimulating factor (GM-CSF) was reported to be a major cause of morbidity in patients infected with highly pathogenic human coronaviruses (SARS-CoV and MERS-CoV) (Channappanavar & Perlman, 2017). In a recent study conducted with 150 patients from Wuhan, China, elevated cytokine levels of IL-6 in patients with COVID-19 was suggested to be the underlying cause of adverse clinical outcomes (Ruan et al., 2020). Many of the recent studies have suggested that cytokine storm, composed of an array of cytokines including IL-1, -2, -6, -7, -8, -10, -12, -17, -18; tumor necrosis factor-α (TNF-α), IFN-γ, granulocyte colony-stimulating factor (G-CSF); GM-CSF, and monocyte chemoattractant protein-1 (MCP-1), is the underlying cause of severe COVID-19 immunopathology (Hadjadj et al., 2020; Lucas et al., 2020; McElvaney et al., 2020; Nile et al., 2020; Ronit et al., 2021).
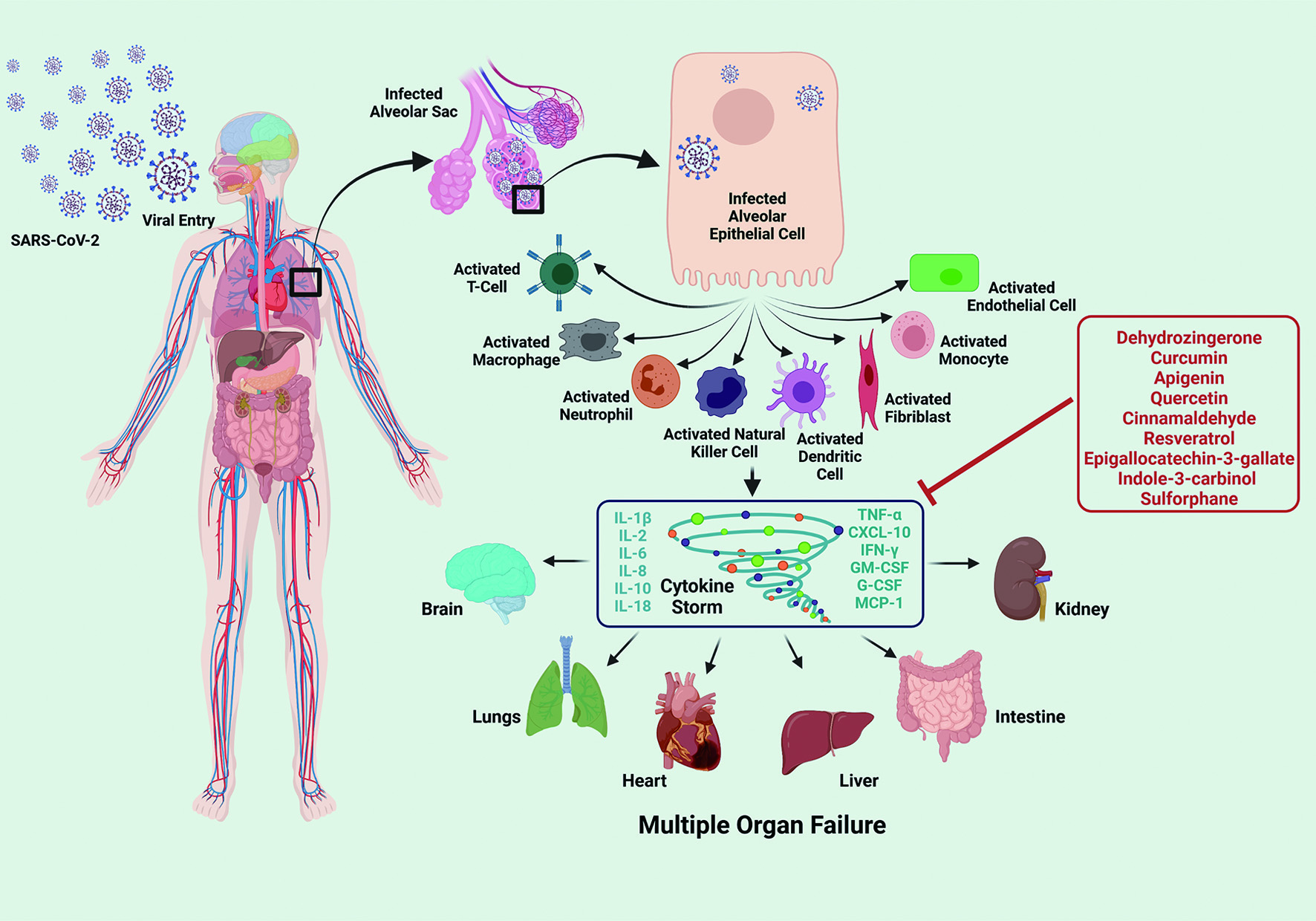 Click for large image | Figure 4. Lead Phytochemicals that can Targeting Pathological Host Response. Upon the entry into the human body, SARS-CoV-2 replicates within the human host cells, especially within the alveolar epithelial cells, as the route of entry is via the respiratory tract. Further, it triggers the immune response via activating the immune cells such as T-cell, alveolar macrophages, neutrophils, natural killer cells, dendritic cells, fibroblasts. monocytes and endothelial cells. These activated immune cells potentiates a cascade of signaling events inducing the release of varied cytokines such as IL-1β/2/6/8/10/18, TNF-α, IFN-γ, G-CSF, GM-CSF, CXCL-10 and MCP-1, producing CRS and cytokine storm, which can lead to multiple organ failure, affecting the major human organs including brain, lungs, liver, heart, intestine and kidney. Phytochemicals that have therapeutic or preventive potential to target pathological CRS and/or cytokine storm are boxed and listed in red. |
Dehydrozingerone, a phenolic compound found in Zingiber officinale (ginger) and a structural-half analog of curcumin, was shown to exert significant inhibitory effect on LPS induced cytokine storm in cultured macrophage cells (Tirunavalli et al., 2021). Many known dietary polyphenols such as curcumin, apigenin, quercetin, cinnamaldehyde, resveratrol, and epigallocatechin-3-gallate; alone and in combinations, have been reported to have significantly reduced the levels of pro-inflammatory cytokines in vitro and in vivo (Giovinazzo and Grieco, 2015; John et al., 2011; Scalbert et al., 2005; Tsao, 2010). Bioactive metabolites Indole-3-carbinol and sulforaphanes present in cruciferous vegetables have also been demonstrated to have a moderate inhibitory effect on LPS-induced inflammatory response in cultured cells (Guo et al., 2012; Jiang et al., 2013). Such phytochemicals can serve as potential lead compounds for the synthesis of effective therapeutics, which could reduce the cytokine storm.
| 7. Perspective and conclusion | ▴Top |
COVID-19 has evolved to be a pandemic with global concern since 2020, which brought the world to a standstill. With the novel vaccine strategies, we are slowly emerging out of the abysmal crisis. However, a critical concern is that an effective therapeutic agent and/or a potential preventive strategy is still elusive. This can be primarily attributed to the enhanced pace of infection and lack of sufficient time for deeper research and validation. During the initial phases of the pandemic, the researchers had come up with the notion of repurposing the existing drugs with antiviral activity, most of which were FDA approved and used against various human diseases. However, most of them, if not all, failed to prove their efficacy in preventing or treating COVID-19 infections in real-world situation. Thus, the need for the hour is to develop novel therapeutic agents which can effectively target the COVID-19 infection in humans. The mechanistic role of phytochemicals discussed here needs to be explored further and experimentally validated so that more potent molecules can be developed. It is quite encouraging to note that the limited examples of phytochemicals discussed here provide evidence that they can serve as critical lead compounds for the development of novel therapeutics for treating and preventing COVID-19 infections or similar pathogens that might emerge in the future.
Acknowledgments
The funding supports from the Stephenson Cancer Center and National Institutes of Health grant (grant no. GM103639) are greatfully acknowledged. Computational services were supported by the National Institute of General Medical Sciences P20 (grant no. GM103639) and the National Cancer Insti-tute of the National Institutes of Health (grant no. P30CA225520).
Conflict of interest
The authors declare that there is no conflict of interest.
| References | ▴Top |